Crotonylation of IDH1 alleviates MASLD progression by enhancing the TCA cycle
- PMID: 40858572
- PMCID: PMC12381242
- DOI: 10.1038/s41467-025-62731-9
Crotonylation of IDH1 alleviates MASLD progression by enhancing the TCA cycle
Abstract
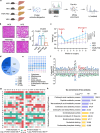
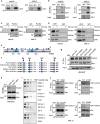
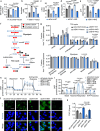
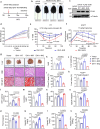
Metabolic dysfunction-associated steatotic liver disease (MASLD), potentially ameliorated by bariatric-metabolic surgery, remains a global health concern in the absence of approved drugs. Protein post-translational modifications (PTMs) are crucial for MASLD. However, the functional significance of lysine crotonylation (Kcr) remains unclear. We aimed to investigate the mechanisms by Kcr-regulated IDH1 in the tricarboxylic acid (TCA) cycle and MASLD development. Herein, we reported a quantitative proteomics analysis of global crotonylome upon MASLD and Post-bariatric. Specifically, decreases in K58cr, K151cr, K212cr and K345cr of IDH1 upon MASLD were observed. PCAF and SIRT7 dynamically regulated the IDH1 Kcr. Abolishment of IDH1 Kcr impaired TCA cycle by decreasing IDH1 enzymatic activity. Male mice with liver-specific expression of crotonylation-mimic mutants of IDH1 were resistant to HFD-induced obesity, insulin resistance, glucose intolerance and MASLD. Our findings unravel the mechanisms of IDH1 Kcr and indicate that targeting PCAF/SIRT7-IDH1 Kcr and metabolites may be a promising strategy for MASLD therapy.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
-
- Powell, E. E., Wong, V. W.-S. & Rinella, M. Non-alcoholic fatty liver disease. Lancet397, 2212–2224 (2021). - PubMed
-
- Rinella, M. E. et al. A multisociety Delphi consensus statement on new fatty liver disease nomenclature. J. Hepatol.79, 1542–1556 (2023). - PubMed
-
- Quek, J. et al. Global prevalence of non-alcoholic fatty liver disease and non-alcoholic steatohepatitis in the overweight and obese population: a systematic review and meta-analysis. Lancet Gastroenterol. Hepatol.8, 20–30 (2023). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

