Programmable epigenome editing by transient delivery of CRISPR epigenome editor ribonucleoproteins
- PMID: 40858609
- PMCID: PMC12381050
- DOI: 10.1038/s41467-025-63167-x
Programmable epigenome editing by transient delivery of CRISPR epigenome editor ribonucleoproteins
Abstract
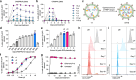
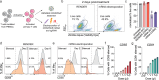
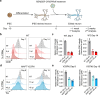
Programmable epigenome editors modify gene expression in mammalian cells by altering the local chromatin environment at target loci without inducing DNA breaks. However, the large size of CRISPR-based epigenome editors poses a challenge to their broad use in biomedical research and as future therapies. Here, we present Robust ENveloped Delivery of Epigenome-editor Ribonucleoproteins (RENDER) for transiently delivering programmable epigenetic repressors (CRISPRi, DNMT3A-3L-dCas9, CRISPRoff) and activator (TET1-dCas9) as ribonucleoprotein complexes into human cells to modulate gene expression. After rational engineering, we show that RENDER induces durable epigenetic silencing of endogenous genes across various human cell types, including primary T cells. Additionally, we apply RENDER to epigenetically repress endogenous genes in human stem cell-derived neurons, including the reduction of the neurodegenerative disease associated V337M-mutated Tau protein. Together, our RENDER platform advances the delivery of CRISPR-based epigenome editors into human cells, broadening the use of epigenome editing in fundamental research and therapeutic applications.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: J.K.N. and D.X. have submitted a patent application to U.S. Provisional Patent Office related to the delivery of CRISPR components, as part of this work (application No. 63/686,584). J.K.N. is an inventor of patents related to the CRISPRoff/on technologies, filed by The Regents of the University of California. B.R.C. is a founder of Tenaya Therapeutics and holds equity in the company. The remaining authors declare no competing interests.
Figures


Update of
-
Programmable epigenome editing by transient delivery of CRISPR epigenome editor ribonucleoproteins.bioRxiv [Preprint]. 2024 Nov 28:2024.11.26.625496. doi: 10.1101/2024.11.26.625496. bioRxiv. 2024. Update in: Nat Commun. 2025 Aug 26;16(1):7948. doi: 10.1038/s41467-025-63167-x. PMID: 39651312 Free PMC article. Updated. Preprint.
References
-
- Villiger, L. et al. CRISPR technologies for genome, epigenome and transcriptome editing. Nat. Rev. Mol. Cell Biol.25, 464–487 (2024). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

