Integrative microbiome- and metatranscriptome-based analyses reveal diagnostic biomarkers for peri-implantitis
- PMID: 40858628
- PMCID: PMC12381052
- DOI: 10.1038/s41522-025-00807-6
Integrative microbiome- and metatranscriptome-based analyses reveal diagnostic biomarkers for peri-implantitis
Abstract
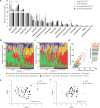
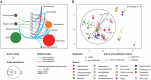
Peri-implantitis is a severe biofilm-associated infection affecting millions worldwide. This cross-sectional study aimed to identify taxonomic and functional biomarkers that reliably indicate peri-implantitis by utilizing paired data from full length 16S rRNA gene amplicon sequencing (full-16S) and metatranscriptomics (RNAseq) in 48 biofilm samples from 32 patients. Both full-16S and RNAseq analyses revealed significant differences between healthy and peri-implantitis samples, with a shift toward anaerobic Gram-negative bacteria in peri-implantitis. Metatranscriptomics identified enzymatic activities and metabolic pathways associated with peri-implantitis and uncovered complex peri-implant biofilm ecology related to amino acid metabolism. Integrating taxonomic and functional data enhanced predictive accuracy (AUC = 0.85) and revealed diagnostic biomarkers including health-associated Streptococcus and Rothia species and peri-implantitis-associated enzymes (urocanate hydratase, tripeptide aminopeptidase, NADH:ubiquinone reductase, phosphoenolpyruvate carboxykinase and polyribonucleotide nucleotidyltransferase). Thus, biofilm profiling at taxonomic and functional levels provides highly predictive disease biomarkers, laying the foundation for novel diagnostic and personalized treatment approaches for peri-implant disease.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






References
-
- Berglundh, T. et al. Peri-implant diseases and conditions: consensus report of workgroup 4 of the 2017 world workshop on the classification of periodontal and peri-implant diseases and conditions. J. Clin. Periodontol.45, S286–S291 (2018). - PubMed
-
- Schwarz, F., Derks, J., Monje, A. & Wang, H. L. Peri-implantitis. J. Periodontol.89, S267–S290 (2018). - PubMed
-
- Derks, J. & Tomasi, C. Peri-implant health and disease. A systematic review of current epidemiology. J. Clin. Periodontol.42, S158–S171 (2015). - PubMed
-
- Dreyer, H. et al. Epidemiology and risk factors of peri-implantitis: a systematic review. J. Periodontal Res.53, 657–681 (2018). - PubMed
MeSH terms
Substances
Grants and funding
- SFB/TRR-298-SIIRI - Project-ID 426335750/Deutsche Forschungsgemeinschaft (German Research Foundation)
- Germany's Excellence Strategy - EXC 2155 - project number 390874280/Deutsche Forschungsgemeinschaft (German Research Foundation)
- SFB/TRR-298-SIIRI - Project-ID 426335750/Deutsche Forschungsgemeinschaft (German Research Foundation)
- SFB/TRR-298-SIIRI - Project-ID 426335750/Deutsche Forschungsgemeinschaft (German Research Foundation)
- SFB/TRR-298-SIIRI - Project-ID 426335750/Deutsche Forschungsgemeinschaft (German Research Foundation)
LinkOut - more resources
Full Text Sources

