EMT-ciliary signaling in quasi-mesenchymal-stem-like cells drives therapeutic resistance and is a druggable vulnerability in triple-negative breast cancer
- PMID: 40859055
- PMCID: PMC12514032
- DOI: 10.1038/s44321-025-00289-1
EMT-ciliary signaling in quasi-mesenchymal-stem-like cells drives therapeutic resistance and is a druggable vulnerability in triple-negative breast cancer
Abstract
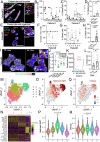
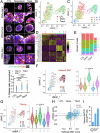
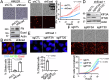
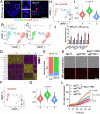
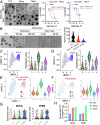
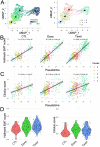
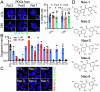
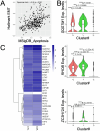
Cancer therapeutic resistance is mediated, in part, by phenotypic heterogeneity and the plasticity of tumor cells, the latter being enabled by epithelial-mesenchymal transition (EMT). However, EMT in human cancer therapeutic response remains poorly understood. We developed patient-derived organoids (PDOs) from human triple-negative breast cancer (TNBC) and investigated their response to chemotherapy. We found that chemotherapy treatment kills the bulk of tumor cells in PDOs, but there is selective survival of malignant cells that had activated an EMT program, entered a quasi-mesenchymal, stem cell-like state and display primary cilia. We developed a family of small-molecule inhibitors of ciliogenesis and show that treatment with these inhibitors, or genetic ablation of primary cilia, is sufficient to suppress this chemoresistance via NFκB-induced cell death. We conclude that an EMT-ciliary signaling axis induces chemoresistance in quasi-mesenchymal ciliated stem-like cells to help tumors evade chemotherapy and represents a druggable vulnerability in human TNBC.
Keywords: EMT; Primary cilia; Therapeutic Resistance; Triple-Negative Breast Cancer.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. A Patient application #EP24199133.0 named INHIBITOR OF CILIOGENESIS FOR USE IN A METHOD OF PREVENTING THERAPEUTIC RESISTANCE IN CANCER was filed by Inserm Transfert.
Figures


References
-
- Bardia A, Hurvitz SA, Tolaney SM, Loirat D, Punie K, Oliveira M, Brufsky A, Sardesai SD, Kalinsky K, Zelnak AB et al (2021) Sacituzumab govitecan in metastatic triple-negative breast cancer. N Engl J Med 384:1529–1541 - PubMed
-
- Biehs B, Dijkgraaf GJP, Piskol R, Alicke B, Boumahdi S, Peale F, Gould SE, de Sauvage FJ (2018) A cell identity switch allows residual BCC to survive Hedgehog pathway inhibition. Nature 562:429–433 - PubMed
-
- Bonnefoi H, Litiere S, Piccart M, MacGrogan G, Fumoleau P, Brain E, Petit T, Rouanet P, Jassem J, Moldovan C et al (2014) Pathological complete response after neoadjuvant chemotherapy is an independent predictive factor irrespective of simplified breast cancer intrinsic subtypes: a landmark and two-step approach analyses from the EORTC 10994/BIG 1-00 phase III trial. Ann Oncol 25:1128–1136 - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

