Dominant negative ATP5F1A variants disrupt oxidative phosphorylation causing neurological disorders
- PMID: 40859057
- PMCID: PMC12514044
- DOI: 10.1038/s44321-025-00290-8
Dominant negative ATP5F1A variants disrupt oxidative phosphorylation causing neurological disorders
Abstract
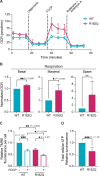
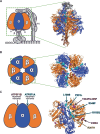
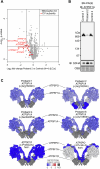
ATP5F1A encodes the α-subunit of complex V of the respiratory chain, which is responsible for mitochondrial ATP synthesis. We describe 6 probands with heterozygous de novo missense ATP5F1A variants that presented with developmental delay, intellectual disability, and movement disorders. All variants were located at the contact points between the α- and β-subunits. Functional studies in C. elegans revealed that the variants were damaging via a dominant negative genetic mechanism. Biochemical and proteomics studies of proband-derived cells showed a marked reduction in complex V abundance and activity. Mitochondrial physiology studies revealed increased oxygen consumption, yet decreased mitochondrial membrane potential and ATP levels indicative of uncoupled oxidative phosphorylation as a pathophysiologic mechanism. Our findings contrast with the previously reported ATP5F1A variant, p.Arg207His, indicating a different pathological mechanism. This study expands the phenotypic and genotypic spectrum of ATP5F1A-associated conditions and highlights how functional studies can provide an understanding of the genetic, molecular, and cellular mechanisms of ATP5F1A variants of uncertain significance. With 12 heterozygous individuals now reported, ATP5F1A is the most frequent nuclear genome cause of complex V deficiency.
Keywords: ATP Synthase; ATP5F1A; Complex V; Mitochondriopathy; Oxidative Phosphorylation.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures






Update of
-
Dominant negative ATP5F1A variants disrupt oxidative phosphorylation causing neurological disorders.medRxiv [Preprint]. 2025 Jul 8:2025.07.08.25330848. doi: 10.1101/2025.07.08.25330848. medRxiv. 2025. Update in: EMBO Mol Med. 2025 Oct;17(10):2562-2585. doi: 10.1038/s44321-025-00290-8. PMID: 40672495 Free PMC article. Updated. Preprint.
References
-
- Abrahams JP, Leslie AG, Lutter R, Walker JE (1994) Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature 370:621–628 - PubMed
-
- Bejsovec A, Anderson P (1988) Myosin heavy-chain mutations that disrupt Caenorhabditis elegans thick filament assembly. Genes Dev 2:1307–1317 - PubMed
MeSH terms
Substances
Grants and funding
- U54 NS108251/NS/NINDS NIH HHS/United States
- P40 OD010440/OD/NIH HHS/United States
- U01 HG007709/HG/NHGRI NIH HHS/United States
- U01HG007709/HHS | NIH | National Human Genome Research Institute (NHGRI)
- 2009982/National Health and Medical Research Council investigator grant
- S10 OD028538-01A1/NIH/NCCR
- S10 OD028538/OD/NIH HHS/United States
- 2019-1198 and 2023-1484/Royal Children's Hospital Foundation (RCHF)
- R35 GM152192/GM/NIGMS NIH HHS/United States
- P50 HD103555/HD/NICHD NIH HHS/United States
- R01 GM100756/GM/NIGMS NIH HHS/United States
- 2016030/Medical Research Future Fund Genomics Health Futures Mission
- U54NS108251/HHS | National Institutes of Health (NIH)
- U54 NS078059/NS/NINDS NIH HHS/United States
- U54NS078059/HHS | National Institutes of Health (NIH)
- 2009732/Australian National Medical Research Council
- U01HG007942/HHS | NIH | National Human Genome Research Institute (NHGRI)
- GM100756/HHS | NIH | National Institute of General Medical Sciences (NIGMS)
- U01 HG007942/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

