Genome-wide characterization and expression analysis of the growth-regulating factor family in Mikania micrantha
- PMID: 40859145
- PMCID: PMC12379477
- DOI: 10.1186/s12870-025-07219-w
Genome-wide characterization and expression analysis of the growth-regulating factor family in Mikania micrantha
Abstract
Background: Growth-regulating factors (GRFs) are plant-specific transcription factors involved in growth, development, and stress responses. Mikania micrantha, a highly invasive weed, displays rapid growth and adaptability, yet the GRF gene family in this species remains largely unexplored.
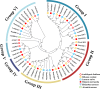
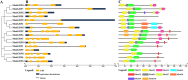
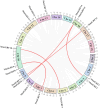
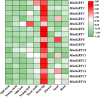
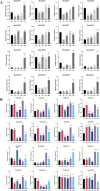
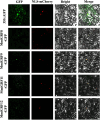
Results: We identified and characterized 16 GRF genes in M. micrantha, analyzing their phylogeny, gene structure, collinearity, and expression patterns. These genes were classified into six subfamilies and showed conserved synteny with GRFs from Arabidopsis thaliana and Asteraceae species, with all gene pairs under purifying selection. Expression profiling revealed high transcript levels of most MmiGRFs in stem tips and young tissues. Hormone treatments demonstrated that MmiGRFs responded differentially to GA3, ABA, IAA, BR, 6-BA, and MeJA, with MmiGRF8 notably upregulated by all treatments. Subcellular localization confirmed nuclear localization of MmiGRF proteins. Additionally, five miR396 members were predicted to target 15 MmiGRFs, with MmiGRF10 containing two distinct sites.
Conclusion: This study provides the first comprehensive analysis of the M. micrantha GRF gene family, revealing their potential roles in rapid growth and environmental adaptation. These findings offer molecular insights into the species' invasive capacity and support future strategies for its control.
Keywords: Mikania micrantha; Expression analysis; Growth and development; Growth-regulating factors.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
- 31872670/National Natural Science Foundation of China
- 31872670/National Natural Science Foundation of China
- 2021A1515010911/Guangdong Basic and Applied Basic Research Foundation
- 2021A1515010911/Guangdong Basic and Applied Basic Research Foundation
- 202206010107/Science and Technology Projects in Guangzhou
LinkOut - more resources
Full Text Sources

