Multi-omics dissection of metabolic and transcriptional regulation underlying fruit maturation in Panax ginseng
- PMID: 40859177
- PMCID: PMC12379430
- DOI: 10.1186/s12870-025-07221-2
Multi-omics dissection of metabolic and transcriptional regulation underlying fruit maturation in Panax ginseng
Abstract
Background: Panax ginseng is a perennial plant valued for its medicinal and nutritional properties. Its fruit contains a variety of bioactive compounds such as ginsenosides, flavonoids, phenolic acids, and anthocyanins. However, the regulatory mechanisms underlying the accumulation of these compounds during fruit development remain largely unexplored.
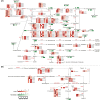
Results: We performed integrated metabolomic and transcriptomic analyses across four developmental stages of ginseng fruit. Metabolite profiling revealed stage-specific accumulation patterns of ginsenosides and phenolics with biphasic trends, and increasing levels of flavonoids and anthocyanins during maturation. We constructed a metabolic and gene expression atlas covering primary metabolism (carbon, amino acids, nitrogen), secondary metabolism (flavonoids, terpenoids), and hormone signaling pathways (abscisic acid, gibberellin, brassinosteroids). Key structural genes and transcription factors, including MYB, bHLH, and ERF families, were found to coordinate stage-specific metabolic shifts. Weighted gene co-expression network analysis revealed metabolite-linked gene modules that delineate regulatory relationships.
Conclusions: This study provides a comprehensive molecular framework of fruit development in P. ginseng, highlighting coordinated transcriptional regulation and metabolic reprogramming. These insights contribute to our understanding of developmental regulation in medicinal plants and lay the groundwork for metabolic engineering strategies aimed at enhancing nutritional quality and bioactive compound production.
Keywords: Panax ginseng; Fruit development; Multi-omics integration; Primary metabolism; Secondary metabolism; Transcription factors.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures







References
-
- Méteignier L-V, Nützmann H-W, Papon N, Osbourn A, Courdavault V. Emerging mechanistic insights into the regulation of specialized metabolism in plants. Nat Plants. 2023;9(1):22–30. - PubMed
-
- Kim Y-J, Joo SC, Shi J, Hu C, Quan S, Hu J, Sukweenadhi J, Mohanan P, Yang D-C, Zhang D. Metabolic dynamics and physiological adaptation of Panax ginseng during development. Plant Cell Rep. 2018;37(3):393–410. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

