All-in-one Biocomputing Nanoagents with Multilayered Transformable Architecture based on DNA Interfaces
- PMID: 40860151
- PMCID: PMC12374668
- DOI: 10.7150/thno.113059
All-in-one Biocomputing Nanoagents with Multilayered Transformable Architecture based on DNA Interfaces
Abstract
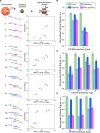
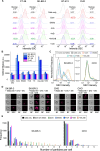
The pathogen diversity to infiltrate the host organism highlights the demand for equally sophisticated mechanisms for their prevention. The development of "intelligent" agents with molecular logic capabilities are of great hope, but their full theranostic potential has yet to be realized. Methods: The original concept of nanoagents based on "Biocomputing based on particle disassembly" technology has been extended to nucleic acids (NAs) interfaces and inputs. By exploiting the unique properties of NAs, we designed nanostructures that can implement all basic single- and dual-input logic gates on a unified nanoparticle platform through DNA strand displacement triggered by oligonucleotide inputs. Performance of nanostructures was investigated across various output signal detection formats including specific interaction with nanosized objects and targeting cells. Results: Here, we demonstrate autonomous theranostic biocomputing agents based on nanoparticles and DNA interfaces ("DNA-transformers") capable of executing a functionally complete set of Boolean logic gates (YES, NOT, AND, and OR) within a single all-in-one particle structure. Each DNA-transformer is constructed through a multi-layered self-assembly of nanoparticles via DNA-interfaces. The route of the agent's disassembly induced by the particular combination of the specific ssDNA inputs determines the agents' ability to produce the programmed outputs compatible with theranostic applications such as specific targeting of HER2/neu-positive cancer cells. Conclusions: The developed all-in-one DNA-based nanoagents represent a significant advancement in molecular logic devices, establishing a versatile platform for smart nanoagents equally suitable for diagnostic and therapeutic applications.
Keywords: DNA interfaces; HER2/neu cell targeting; biocomputing; smart materials; strand displacement.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures







Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Management of urinary stones by experts in stone disease (ESD 2025).Arch Ital Urol Androl. 2025 Jun 30;97(2):14085. doi: 10.4081/aiua.2025.14085. Epub 2025 Jun 30. Arch Ital Urol Androl. 2025. PMID: 40583613 Review.
-
Biocomputing systems based on carbon materials and DNA.Nanoscale. 2025 Aug 21;17(33):19055-19069. doi: 10.1039/d5nr02383a. Nanoscale. 2025. PMID: 40758426 Review.
-
MarkVCID cerebral small vessel consortium: I. Enrollment, clinical, fluid protocols.Alzheimers Dement. 2021 Apr;17(4):704-715. doi: 10.1002/alz.12215. Epub 2021 Jan 21. Alzheimers Dement. 2021. PMID: 33480172 Free PMC article.
-
Comparison of self-administered survey questionnaire responses collected using mobile apps versus other methods.Cochrane Database Syst Rev. 2015 Jul 27;2015(7):MR000042. doi: 10.1002/14651858.MR000042.pub2. Cochrane Database Syst Rev. 2015. PMID: 26212714 Free PMC article.
References
-
- Tregubov AA, Nikitin PI, Nikitin MP. Advanced smart nanomaterials with integrated logic-gating and biocomputing: dawn of theranostic nanorobots. Chem Rev. 2018;118:10294–348. - PubMed
-
- Nikitin MP, Shipunova VO, Deyev SM, Nikitin PI. Biocomputing based on particle disassembly. Nat Nanotechnol. 2014;9:716–22. - PubMed
-
- Nikitin MP. Non-complementary strand commutation as a fundamental alternative for information processing by DNA and gene regulation. Nat Chem. 2023;15:70–82. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

