TSG6 promotes epithelial-mesenchymal transition and tumor-associated macrophage polarization through Smad2/3 and MAPK signaling by facilitating TSG6-CD44-TGFβR1 or EGFR complex formation
- PMID: 40860188
- PMCID: PMC12374808
- DOI: 10.7150/ijbs.115097
TSG6 promotes epithelial-mesenchymal transition and tumor-associated macrophage polarization through Smad2/3 and MAPK signaling by facilitating TSG6-CD44-TGFβR1 or EGFR complex formation
Abstract
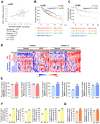
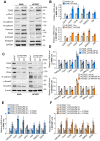
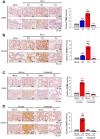
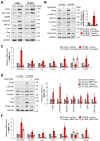
TSG6 is highly expressed during PLK1-induced epithelial-mesenchymal transition (EMT). However, the role of TSG6 in the tumor microenvironment (TME) remains poorly understood. We investigate the function and regulatory mechanisms of TSG6 in immune plasticity within the TME of lung adenocarcinoma (LUAD). The simultaneous high expression of TSG6 and PLK1 in LUAD patients was associated with lower survival rates. TSG6 and CD44 were markedly upregulated during EMT driven by TGF-b or active PLK1 in A549 and HCC827 cells. TSG6 treatment enhanced EMT by increasing N-cadherin and phosphorylated Smad2 levels. TSG6 depletion blocked the effects, which was restored upon TSG6 retreatment. Additionally, TSG6 treatment induced polarization of THP-1 monocytes into M2d tumor-associated macrophages (TAMs). In cocultures of THP-1 monocytes with A549 cells expressing TSG6, M2d-inducing factors in A549 cells and M2d markers in THP-1 cells were upregulated. Immunoprecipitation showed that TSG6 binds CD44, enhancing CD44's interaction with TGFbR or EGFR. In TSG6-treated LUAD cells, both total CD44 and its cleaved intracellular domain increased by activating TGFβR1-Smad2/3 and MAPK-ERK1/2-AP-1 pathways. Thus, TSG6 promotes EMT and M2d-TAMs polarization by activating TGFβR1/Smad and MAPK/ERK pathway through direct interaction between CD44 and TGFβR1 or EGFR.
Keywords: CD44; TSG6; invasiveness; tumor-associated macrophages.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

