Methylation-mediated LncRNA CRAT40 promotes colorectal cancer progression by recruiting YBX1 to initiate RelA transcription
- PMID: 40860198
- PMCID: PMC12374820
- DOI: 10.7150/ijbs.105629
Methylation-mediated LncRNA CRAT40 promotes colorectal cancer progression by recruiting YBX1 to initiate RelA transcription
Abstract
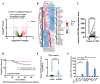
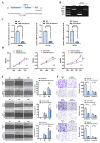
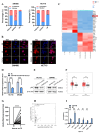
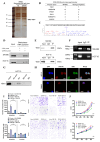
Colorectal cancer (CRC) remains a leading cause of cancer-related mortality worldwide. Long noncoding RNAs (lncRNAs) have emerged as crucial regulators in the initiation and progression of various malignancies, including CRC. In this study, we found that lnc-CRAT40 was upregulated in CRC and associated with poor prognosis following CRC resection. Functional assays revealed that elevated lnc-CRAT40 expression promotes tumor cell proliferation and metastasis both in vitro and in vivo. The modification of N6-methyladenosine, driven by METTL3, was essential for the stability of lnc-CRAT40, which may partially contribute to the upregulation of lnc-CRAT40. Mechanistically, lnc-CRAT40 directly interacted with Y-box binding protein 1 (YBX1) and recruits it to the RelA promoter, thereby activating NF-κB signaling, which in turn drives CRC proliferation and metastatic potential. These findings provide novel insights into the molecular mechanisms underlying CRC progression and highlight lnc-CRAT40 as a potential prognostic biomarker and therapeutic target.
Keywords: METTL3; YBX1; colorectal cancer; long non-coding RNA.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures




References
-
- Global Burden of Disease 2019 Cancer Collaboration; Kocarnik JM, Compton K. et al. Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life Years for 29 Cancer Groups From 2010 to 2019: A Systematic Analysis for the Global Burden of Disease Study 2019. JAMA Oncol. 2022;8:420–444. - PMC - PubMed
-
- Biller LH, Schrag D. Diagnosis and Treatment of Metastatic Colorectal Cancer: A Review. JAMA. 2021;325:669–685. - PubMed
-
- Adnane S, Marino A, Leucci E. LncRNAs in human cancers: signal from noise. Trends Cell Biol. 2022;32:565–573. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

