Terrier: a deep learning repeat classifier
- PMID: 40862518
- PMCID: PMC12381760
- DOI: 10.1093/bib/bbaf442
Terrier: a deep learning repeat classifier
Abstract
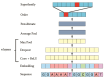
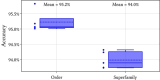
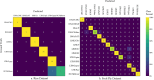
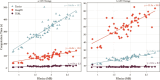
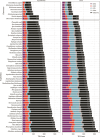
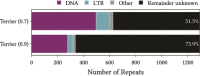
Repetitive DNA sequences underpin genome architecture and evolutionary processes, yet they remain challenging to classify accurately. Terrier is a deep learning model designed to overcome these challenges by classifying repetitive DNA sequences using a publicly available, curated repeat sequence library trained under the RepeatMasker schema. Poor representation of taxa within repeat databases often limits the classification accuracy and reproducibility of current repeat annotation methods, limiting our understanding of repeat evolution and function. Terrier overcomes these challenges by leveraging deep learning for improved accuracy. Trained on Repbase, which includes over 100,000 repeat families-four times more than Dfam-Terrier maps 97.1% of Repbase sequences to RepeatMasker categories, offering the most comprehensive classification system available. When benchmarked against DeepTE, TERL, and TEclass2 in model organisms (rice, fruit flies, humans, and mice), Terrier achieved superior accuracy while classifying a broader range of sequences. Further validation in non-model amphibian, flatworm, and Northern krill genomes highlights its effectiveness in improving classification in non-model species, facilitating research on repeat-driven evolution, genomic instability, and phenotypic variation.
Keywords: DNA sequence classification; Northern krill; amphibians; deep learning; flatworms; transposable elements (TEs).
© The Author(s) 2025. Published by Oxford University Press.
Conflict of interest statement
No competing interest is declared.
Figures



References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

