Autoencoder-Assisted Stacked Ensemble Learning for Lymphoma Subtype Classification: A Hybrid Deep Learning and Machine Learning Approach
- PMID: 40863882
- PMCID: PMC12389832
- DOI: 10.3390/tomography11080091
Autoencoder-Assisted Stacked Ensemble Learning for Lymphoma Subtype Classification: A Hybrid Deep Learning and Machine Learning Approach
Abstract
Background: Accurate subtype identification of lymphoma cancer is crucial for effective diagnosis and treatment planning. Although standard deep learning algorithms have demonstrated robustness, they are still prone to overfitting and limited generalization, necessitating more reliable and robust methods.
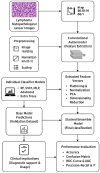
Objectives: This study presents an autoencoder-augmented stacked ensemble learning (SEL) framework integrating deep feature extraction (DFE) and ensembles of machine learning classifiers to improve lymphoma subtype identification.
Methods: Convolutional autoencoder (CAE) was utilized to obtain high-level feature representations of histopathological images, followed by dimensionality reduction via Principal Component Analysis (PCA). Various models were utilized for classifying extracted features, i.e., Random Forest (RF), Support Vector Machine (SVM), Multi-Layer Perceptron (MLP), AdaBoost, and Extra Trees classifiers. A Gradient Boosting Machine (GBM) meta-classifier was utilized in an SEL approach to further fine-tune final predictions.
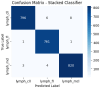
Results: All the models were tested using accuracy, area under the curve (AUC), and Average Precision (AP) metrics. The stacked ensemble classifier performed better than all the individual models with a 99.04% accuracy, 0.9998 AUC, and 0.9996 AP, far exceeding what regular deep learning (DL) methods would achieve. Of standalone classifiers, MLP (97.71% accuracy, 0.9986 AUC, 0.9973 AP) and Random Forest (96.71% accuracy, 0.9977 AUC, 0.9953 AP) provided the best prediction performance, while AdaBoost was the poorest performer (68.25% accuracy, 0.8194 AUC, 0.6424 AP). PCA and t-SNE plots confirmed that DFE effectively enhances class discrimination.
Conclusion: This study demonstrates a highly accurate and reliable approach to lymphoma classification by using autoencoder-assisted ensemble learning, reducing the misclassification rate and significantly enhancing the accuracy of diagnosis. AI-based models are designed to assist pathologists by providing interpretable outputs such as class probabilities and visualizations (e.g., Grad-CAM), enabling them to understand and validate predictions in the diagnostic workflow. Future studies should enhance computational efficacy and conduct multi-centre validation studies to confirm the model's generalizability on extensive collections of histopathological datasets.
Keywords: autoencoder; deep feature extraction; digital pathology; lymphoma classification; machine learning; stacked ensemble learning.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

