Putative PINK1/Parkin activators lower the threshold for mitophagy by sensitizing cells to mitochondrial stress
- PMID: 40864725
- PMCID: PMC12383277
- DOI: 10.1126/sciadv.ady0240
Putative PINK1/Parkin activators lower the threshold for mitophagy by sensitizing cells to mitochondrial stress
Abstract
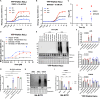
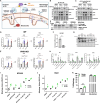
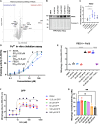
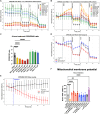
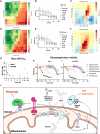
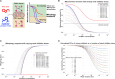
The PINK1/Parkin pathway targets damaged mitochondria for degradation via mitophagy. Genetic evidence implicates impaired mitophagy in Parkinson's disease, making its pharmacological enhancement a promising therapeutic strategy. Here, we characterize two mitophagy activators: a novel Parkin activator, FB231, and the reported PINK1 activator MTK458. Both compounds lower the threshold for mitochondrial toxins to induce PINK1/Parkin-mediated mitophagy. However, global proteomics revealed that FB231 and MTK458 independently induce mild mitochondrial stress, resulting in impaired mitochondrial function and activation of the integrated stress response, effects that result from PINK1/Parkin-independent off-target activities. We find that these compounds impair mitochondria by distinct mechanisms and synergistically decrease mitochondrial function and cell viability in combination with classical mitochondrial toxins. Our findings support a model whereby weak or "silent" mitochondrial toxins potentiate other mitochondrial stressors, enhancing PINK1/Parkin-mediated mitophagy. These insights highlight important considerations for therapeutic strategies targeting mitophagy activation in Parkinson's disease.
Figures



References
-
- Moon H. E., Yoon S. H., Hur Y. S., Park H. W., Ha J. Y., Kim K. H., Shim J. H., Yoo S. H., Son J. H., Paek S. L., Kim I. K., Hwang J. H., Kim D. G., Kim H. J., Jeon B. S., Park S. S., Paek S. H., Mitochondrial dysfunction of immortalized human adipose tissue-derived mesenchymal stromal cells from patients with Parkinson’s disease. Exp. Neurobiol. 22, 283–300 (2013). - PMC - PubMed
-
- Tanner C. M., Kamel F., Ross G. W., Hoppin J. A., Goldman S. M., Korell M., Marras C., Bhudhikanok G. S., Kasten M., Chade A. R., Comyns K., Richards M. B., Meng C., Priestley B., Fernandez H. H., Cambi F., Umbach D. M., Blair A., Sandler D. P., Langston J. W., Rotenone, paraquat, and Parkinson’s disease. Environ. Health Perspect. 119, 866–872 (2011). - PMC - PubMed
-
- Flones I. H., Toker L., Sandnes D. A., Castelli M., Mostafavi S., Lura N., Shadad O., Fernandez-Vizarra E., Painous C., Perez-Soriano A., Compta Y., Molina-Porcel L., Alves G., Tysnes O. B., Dolle C., Nido G. S., Tzoulis C., Mitochondrial complex I deficiency stratifies idiopathic Parkinson’s disease. Nat. Commun. 15, 3631 (2024). - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

