Functional Characterization of Two Glutamate Dehydrogenase Genes in Bacillus altitudinis AS19 and Optimization of Soluble Recombinant Expression
- PMID: 40864757
- PMCID: PMC12384151
- DOI: 10.3390/cimb47080603
Functional Characterization of Two Glutamate Dehydrogenase Genes in Bacillus altitudinis AS19 and Optimization of Soluble Recombinant Expression
Abstract
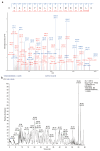
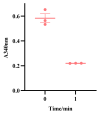
Glutamate dehydrogenase (GDH) is ubiquitous in organisms and crucial for amino acid metabolism, energy production, and redox balance. The gdhA and gudB genes encoding GDH were identified in Bacillus altitudinis AS19 and shown to be regulated by iron. However, their functions remain unclear. In this study, gdhA and gudB were analyzed using bioinformatics tools, such as MEGA, Expasy, and SWISS-MODEL, expressed with a prokaryotic expression system, and the induction conditions were optimized to increase the yield of soluble proteins. Phylogenetic analysis revealed that GDH is evolutionarily conserved within the genus Bacillus. GdhA and GudB were identified as hydrophobic proteins, not secreted or membrane proteins. Their structures were primarily composed of irregular coils and α-helices. SWISS-MODEL predicts GdhA to be an NADP-specific GDH, whereas GudB is an NAD-specific GDH. SDS-PAGE analysis showed that GdhA was expressed as a soluble protein after induction with 0.2 mmol/L IPTG at 24 °C for 16 h. GudB was expressed as a soluble protein after induction with 0.1 mmol/L IPTG at 16 °C for 12 h. The proteins were confirmed by Western blot and mass spectrometry. The enzyme activity of recombinant GdhA was 62.7 U/mg with NADPH as the coenzyme. This study provides a foundation for uncovering the functions of two GDHs of B. altitudinis AS19.
Keywords: Bacillus; bioinformatics; glutamate dehydrogenase; soluble expression.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





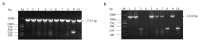



Similar articles
-
A promiscuous Bcd amino acid dehydrogenase promotes biofilm development in Bacillus subtilis.NPJ Biofilms Microbiomes. 2025 Jun 21;11(1):112. doi: 10.1038/s41522-025-00750-6. NPJ Biofilms Microbiomes. 2025. PMID: 40541930 Free PMC article.
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Glutamate dehydrogenase from Pantoea ananatis: A new bacterial enzyme with dual coenzyme specificity.PLoS One. 2025 Aug 19;20(8):e0328289. doi: 10.1371/journal.pone.0328289. eCollection 2025. PLoS One. 2025. PMID: 40828780 Free PMC article.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
References
-
- Ye H.-Y., Hou W., Ruan B.-F. The basic characteristics of glutamate dehydrogenase and its inhibitors. Drug Inf. 2019;8:195–201.
LinkOut - more resources
Full Text Sources

