Splicing QTL mapping in stimulated macrophages associates low-usage splice junctions with immune-mediated disease risk
- PMID: 40866368
- PMCID: PMC12391537
- DOI: 10.1038/s41467-025-61669-2
Splicing QTL mapping in stimulated macrophages associates low-usage splice junctions with immune-mediated disease risk
Abstract
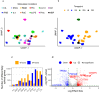
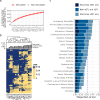
The majority of immune-mediated disease (IMD) risk loci are located in non-coding regions of the genome, making it difficult to decipher their functional effects in relevant physiological contexts. To assess the extent to which alternative splicing contributes to IMD risk, we mapped genetic variants associated with alternative splicing (splicing quantitative trait loci or sQTL) in macrophages exposed to a wide range of environmental stimuli. We found that genes involved in innate immune response pathways undergo extensive differential splicing in response to stimulation and detected significant sQTL effects for over 5734 genes across all stimulation conditions. We colocalised sQTL signals for over 700 genes with IMD-associated risk loci from 22 IMDs with high confidence (PP4 ≥ 0.75). Approximately half of the colocalisations implicate lowly-used splice junctions (mean usage ratio <0.1). Finally, we demonstrate how an inflammatory bowel disease (IBD) risk allele increases the usage of a lowly-used isoform of PTPN2, a negative regulator of inflammation. Together, our findings highlight the role alternative splicing plays in IMD risk, and suggest that lowly-used splicing events significantly contribute to complex disease risk.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: C.A.A. has received research grants or consultancy/speaker fees from Genomics plc, BridgeBio, G.S.K. and AstraZeneca. D.J.G. was an employee of BioMarin and N.I.P. was an employee of G.S.K. at the time the manuscript was submitted. The remaining authors do not declare any competing financial or non-financial interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

