In silico characterization, physiochemical analysis, and antifungal evaluation of the Zea mays PR-1 protein
- PMID: 40866506
- PMCID: PMC12391533
- DOI: 10.1038/s41598-025-16772-1
In silico characterization, physiochemical analysis, and antifungal evaluation of the Zea mays PR-1 protein
Abstract
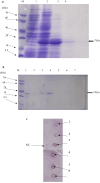
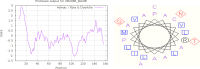
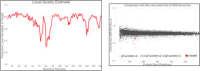
Pathogenesis-related protein 1 (PR-1) is a critical component of plant defense mechanisms, exhibiting significant antifungal activity. This study focused on the PR-1 protein from Zea mays and five other plant species (Triticum aestivum, Hordeum vulgare, Oryza sativa, Avena sativa, and Secale cereale) to explore their physicochemical, structural, and functional characteristics. The PR-1 gene from Z. mays was isolated and expressed in Escherichia coli BL21-DE3 cells, enabling structural characterization. Recombinant expression in E. coli enabled structural characterization and functional studies of the PR-1 protein, laying a foundation for future experiments. Sequence analysis revealed high similarity (65-90%) among PR-1 proteins across species, although physicochemical properties varied based on plant origin. Structural analysis showed a conserved beta-barrel structure surrounded by alpha-helices in all PR-1 proteins. In addition to in silico and structural characterization, antifungal bioassays were performed using the purified recombinant PR-1 protein against Fusarium oxysporum, a major fungal phytopathogen. These assays demonstrated notable antifungal activity, supporting the predicted defense role of PR-1 and laying experimental groundwork for its potential use in developing fungi-resistant transgenic crops.
Keywords: Zea mays; Pathogen resistance; Pathogenesis-related protein; Structural analysis.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures








Similar articles
-
Investigating the efficacy of bioactive compounds from selected plant extracts against Gibberella fujikuroi species complex associated with damping off disease in sweet corn.Sci Rep. 2025 Jul 1;15(1):21712. doi: 10.1038/s41598-025-05979-x. Sci Rep. 2025. PMID: 40594364 Free PMC article.
-
Mycogenic synthesis of silver nanoparticles using endophytic fungi and their characterization, biological activities, including in-silico studies with special reference to Fusarium wilt of tomato.Fungal Biol. 2025 Aug;129(5):101610. doi: 10.1016/j.funbio.2025.101610. Epub 2025 Jun 4. Fungal Biol. 2025. PMID: 40707118
-
Transcriptomic Characterization of Candidate Genes for Fusarium Resistance in Maize (Zea mays L.).Pathogens. 2025 Aug 6;14(8):779. doi: 10.3390/pathogens14080779. Pathogens. 2025. PMID: 40872289 Free PMC article.
-
Antifungal agents for preventing fungal infections in non-neutropenic critically ill patients.Cochrane Database Syst Rev. 2016 Jan 16;2016(1):CD004920. doi: 10.1002/14651858.CD004920.pub3. Cochrane Database Syst Rev. 2016. PMID: 26772902 Free PMC article.
-
Epidemiology of Triazole Resistant Aspergillus fumigatus in Asia: A Systematic Review and Meta-Analysis.Mycoses. 2025 Aug;68(8):e70099. doi: 10.1111/myc.70099. Mycoses. 2025. PMID: 40792442 Review.
References
-
- Dalio, R. J. et al. Hypersensitive response: From NLR pathogen recognition to cell death response. Ann. Appl. Biol.178, 268–280 (2021).
-
- Doughari, J. An overview of plant immunity. J. Plant Pathol. Microbiol.6, 104172 (2015).
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Research Materials

