Cefiderocol-resistant pathogens in German hospital wastewater: a reservoir for multidrug resistance
- PMID: 40866524
- PMCID: PMC12391349
- DOI: 10.1038/s41598-025-17379-2
Cefiderocol-resistant pathogens in German hospital wastewater: a reservoir for multidrug resistance
Abstract
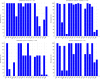
Cefiderocol-resistant bacteria pose a growing concern in both clinical and environmental settings. This study investigates cefiderocol-resistant bacteria in wastewater from six German tertiary care hospitals. A total of 36 samples were analysed using a culture-dependent approach involving cefiderocol pre-enrichment, yielding 97 cefiderocol-resistant isolates-primarily Enterobacter roggenkampii, Klebsiella oxytoca, Serratia marcescens, and Citrobacter farmeri. Most isolates exhibited high minimum inhibitory concentrations against cefiderocol and resistance to multiple antibiotics. Resistance rates were lower for meropenem-vaborbactam (10.3%) and imipenem-relebactam (33.0%), while all isolates remained susceptible to aztreonam-avibactam. Whole-genome sequencing of 79 isolates revealed a diverse resistome, with 78.5% (62/79) carrying carbapenemase genes. Some isolates harbored up to six distinct β-lactamase genes, including combinations of extended-spectrum β-lactamases (ESBLs), AmpC β-lactamases, and one or multiple carbapenemases-such as as blaOXA-48 co-occurring with blaNDM-1 or blaVIM-1. In addition, biocide and heavy metal resistance genes were prevalent, highlighting bacterial adaptation to harsh environments. Plasmid profiling showed significant interspecies variation, with C. farmeri and K. oxytoca displaying the highest plasmid loads. Across all isolates, 38 unique plasmid incompatibility types were detected, 18 of which were species-specific. These findings highlight the multidrug-resistant nature of wastewater-derived pathogens and the importance of monitoring resistance dissemination in healthcare environments.
Keywords: Carbapenemases; Cefiderocol resistance; Multidrug-resistant pathogens; Pandrug-resistant bacteria; Plasmid incompatibility types; Wastewater surveillance.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures

References
-
- WHO Bacterial Priority Pathogens List. Bacterial pathogens of public health importance to guide research, development and strategies to prevent and control antimicrobial resistance. (2024). Available at https://iris.who.int/bitstream/handle/10665/376776/9789240093461-eng.pdf.... - PMC - PubMed
-
- ECDC and WHO. Antimicrobial resistance surveillance in Europe. (2023). Available at https://www.ecdc.europa.eu/sites/default/files/documents/Antimicrobial%2....
-
- Zhanel, G. G. et al. Imipenem-Relebactam and Meropenem-Vaborbactam: two novel Carbapenem-β-Lactamase inhibitor combinations. Drugs78, 65–98 (2018). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

