Maternal stress triggers early-life eczema through fetal mast cell programming
- PMID: 40866704
- PMCID: PMC12488486
- DOI: 10.1038/s41586-025-09419-8
Maternal stress triggers early-life eczema through fetal mast cell programming
Abstract
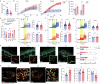
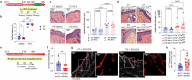
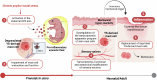
Prenatal stress (PS) is a repeated exposure to aversive situations during pregnancy, including high emotional strain, which is suspected to affect homeostatic systems in infants. Paediatric eczema develops quickly after birth at flexural sites subjected to continuous mechanical constraints1,2. Although epidemiological studies have suggested an association between PS and a higher risk of eczema in children3-6, no causative biological link has yet been identified. Here we show that eczema at birth originates from molecular dysregulations of neuroimmune circuits in utero, triggered by fluctuations in the maternal hypothalamic-pituitary-adrenal axis. We found that offspring of stressed pregnant dams have dysregulated mast cells and skin-projecting neurons and quickly develop eczema in response to harmless mechanical friction. We demonstrated that PS transiently modulates amniotic fluid corticosterone concentrations, which directly alters the activation program of skin mast cells expressing the glucocorticoid receptor Nr3c1 and the adjacent sensory neurons conveying mechanosensation. Therapeutic normalization of maternal corticosterone concentrations or genetic depletion of Mcpt5+ mast cells during stressed gestation prevents fetal immune dysregulation and protects against eczema development after birth. Our findings support a new model in which early-onset paediatric eczema originates from dysregulations in the fetal immune system, caused by fluctuations in maternal glucocorticoids induced by stress.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: N. Gaudenzio is (or was) collaborating, consulting or serving as a member of the scientific advisory board for Genoskin (where he is CSO and a shareholder), Escient pharmaceuticals, Aikium, CEVA, MaxiVAX, Boehringer Ingelheim, Novartis, Sanofi, Allegria and argenx. L.R. is (or was) collaborating or consulting for Neovacs, Novartis, CEVA and argenx. The other authors declare no competing interests.
Figures










References
-
- Langan, S. M., Irvine, A. D. & Weidinger, S. Atopic dermatitis. Lancet396, 345–360 (2020). - PubMed
-
- Bieber, T. et al. Clinical phenotypes and endophenotypes of atopic dermatitis: where are we, and where should we go? J. Allergy Clin. Immunol.139, S58–S64 (2017). - PubMed
-
- Smejda, K. et al. Maternal stress during pregnancy and allergic diseases in children during the first year of life. Respir. Care63, 70–76 (2018). - PubMed
-
- Larsen, A. D. et al. Exposure to psychosocial job strain during pregnancy and odds of asthma and atopic dermatitis among 7-year old children—a prospective cohort study. Scand. J. Work Environ. Health40, 639–648 (2014). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

