Unraveling the Clinical Landscape of RNA Modification Regulators with Multi-Omics Insights in Pan-Cancer
- PMID: 40867324
- PMCID: PMC12384070
- DOI: 10.3390/cancers17162695
Unraveling the Clinical Landscape of RNA Modification Regulators with Multi-Omics Insights in Pan-Cancer
Abstract
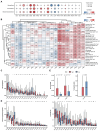
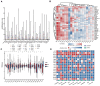
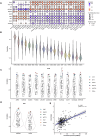
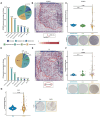
Background/Objectives: Cancer remains a major global health challenge, with RNA modifications increasingly recognized as key regulators of tumor progression. However, integrated pan-cancer analyses across multiple modification types are limited. Methods: We performed a comprehensive analysis of 170 RNA modification-related genes across 33 cancer types, uncovering diverse expression, mutation, and epigenetic patterns. Results: Key regulators such as IGF2BP3, CFI, and ELF3 showed cancer-specific prognostic significance. We developed an RNA Modification Score (RMS) with strong prognostic performance (AUC up to 0.92), correlating with the tumor stage, immune infiltration, and immunotherapy response. High-risk groups exhibited immune checkpoint dysregulation and enriched M1 macrophages in glioblastoma. Drug screening highlighted oncrasin-72 as a potential therapy. Validation via single-cell/spatial transcriptomics and immunohistochemistry confirmed the spatial localization of critical genes like CFI and ELF3. Conclusions: Our study reveals the multifaceted role of RNA modifications in cancer, providing a translational framework for personalized prognosis and therapy in precision oncology.
Keywords: RNA modifications; pan-cancer analysis; precision oncology; prognostic model; tumor microenvironment.
Conflict of interest statement
All the authors declare that they have no conflicts of interest.
Figures


Similar articles
-
Interplay between tumor mutation burden and the tumor microenvironment predicts the prognosis of pan-cancer anti-PD-1/PD-L1 therapy.Front Immunol. 2025 Jul 24;16:1557461. doi: 10.3389/fimmu.2025.1557461. eCollection 2025. Front Immunol. 2025. PMID: 40777041 Free PMC article.
-
Multi-omics analysis unveils the predictive value of IGF2BP3/SPHK1 signaling in cancer stem cells for prognosis and immunotherapeutic response in muscle-invasive bladder cancer.J Transl Med. 2024 Oct 4;22(1):900. doi: 10.1186/s12967-024-05685-8. J Transl Med. 2024. PMID: 39367493 Free PMC article.
-
Comprehensive pan-cancer analysis reveals NTN1 as an immune infiltrate risk factor and its potential prognostic value in SKCM.Sci Rep. 2025 Jan 25;15(1):3223. doi: 10.1038/s41598-025-85444-x. Sci Rep. 2025. PMID: 39863609 Free PMC article.
-
Targeting epigenetic regulators as a promising avenue to overcome cancer therapy resistance.Signal Transduct Target Ther. 2025 Jul 18;10(1):219. doi: 10.1038/s41392-025-02266-z. Signal Transduct Target Ther. 2025. PMID: 40675967 Free PMC article. Review.
-
Systemic treatments for metastatic cutaneous melanoma.Cochrane Database Syst Rev. 2018 Feb 6;2(2):CD011123. doi: 10.1002/14651858.CD011123.pub2. Cochrane Database Syst Rev. 2018. PMID: 29405038 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

