Identification of Two Critical Contact Residues in a Pathogenic Epitope from Tetranectin for Monoclonal Antibody Binding and Preparation of Single-Chain Variable Fragments
- PMID: 40867545
- PMCID: PMC12383646
- DOI: 10.3390/biom15081100
Identification of Two Critical Contact Residues in a Pathogenic Epitope from Tetranectin for Monoclonal Antibody Binding and Preparation of Single-Chain Variable Fragments
Abstract
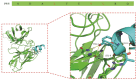
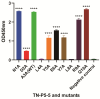
Sepsis is a fetal disease that requires a clear diagnostic biomarker for timely antibiotic treatment. Recent research has identified a pyroptosis-inducing epitope known as P5-5 in tetranectin (TN), a plasma protein produced by monocytes. Previously, we produced a 12F1 monoclonal antibody against the P5-5 and discovered that it could not only diagnose the presence but also monitor the progress of sepsis in the clinic. In the current study, we further investigated the structure site of the P5-5 and the recognition mechanism between the 12F1 mAb and the P5-5 epitope. To this end, 10 amino acids (NDALYEYLRQ) in the P5-5 were individually mutated to alanine, and their binding to the mAb was tested to confirm the most significant antigenic recognition sites. In the meanwhile, the spatial conformation of 12F1 mAb variable regions was modeled, and the molecular recognition mechanisms in detail of the mAb to the P5-5 epitope were further studied by molecular docking. Following epitope prediction and experimental verification, we demonstrated that the motif "DALYEYL" in the epitope sequence position 2-8 of TN-P5-5 is the major binding region for mAb recognition, in which two residues (4L and 8L) were essential for the interaction between the P5-5 epitope and the 12F1 mAb. Therefore, our study greatly narrowed down the previously reported motif from ten to seven amino acids and identified two Leu as critical contact residues. Finally, a single-chain variable fragment (scFv) from the 12F1 hybridoma was constructed, and it was confirmed that the identified motif and residues are prerequisites for the strong binding between P5-5 and 12F1. Altogether, the data of the present work could serve as a theoretic guide for the clinical design of biosynthetic drugs by artificial intelligence to treat sepsis.
Keywords: P5-5; epitope; scFv; sepsis; tetranectin.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Yang X., Pu X., Xu Y., Zhao J., Fang X., Cui J., Deng G., Liu Y., Zhu L., Shao M., et al. A novel prognosis evaluation indicator of patients with sepsis created by integrating six microfluidic-based neutrophil chemotactic migration parameters. Talanta. 2025;281:126801. doi: 10.1016/j.talanta.2024.126801. - DOI - PubMed
-
- Singer M., Deutschman C.S., Seymour C.W., Shankar-Hari M., Annane D., Bauer M., Bellomo R., Bernard G.R., Chiche J.D., Coopersmith C.M., et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3) JAMA. 2016;315:801–810. doi: 10.1001/jama.2016.0287. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

