Piezo1 Channel Activators Yoda1 and Yoda2 in the Context of Red Blood Cells
- PMID: 40867555
- PMCID: PMC12384225
- DOI: 10.3390/biom15081110
Piezo1 Channel Activators Yoda1 and Yoda2 in the Context of Red Blood Cells
Abstract
Piezo1 is a mechanosensitive non-selective cation channel. Genetic alterations of the channel result in a hematologic phenotype named Hereditary Xerocytosis. With Yoda1 and, more recently, Yoda2, compounds to increase the activity of Piezo1 have become available. However, their concrete effect depends on the nano environment of the channel and hence on the cell type. Here we compare the potency of Yoda1 and Yoda2 in red blood cells (RBCs). We investigate the effect of the compounds on direct channel activity using automated patch clamp, as well as the secondary effects of channel activation on signalling molecules and cellular response. In terms of signalling, we investigate the temporal response of the second messenger Ca2+, and in terms of cellular response, the activity of the Gárdos channel. The opening of the Gárdos channel leads to a hyperpolarisation of the RBCs, which is measured by the Macey-Bennekou-Egée (MBE) method. Although the interpretation of the data is not straightforward, we discuss the results in a physiological context and provide recommendations for the use of Yoda1 and Yoda2 to investigate RBCs.
Keywords: MBE-method; Piezo1; Yoda1; Yoda2; automated patch-clamp; erythrocytes; intracellular calcium; live cell imaging; mechanosensitive ion channel; membrane potential.
Conflict of interest statement
The funders had no role in the design of the study; in the collection, analysis, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results. RP, NM, and MGR are employees of Nanion Technologies, the manufacturer of the device used for the acquisition of the data presented in Figure 1 and Figure A1.
Figures


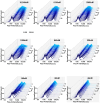


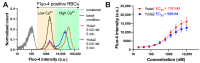

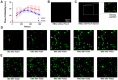


References
-
- Ranade S.S., Qiu Z., Woo S.-H., Hur S.S., Murthy S.E., Cahalan S.M., Xu J., Mathur J., Bandell M., Coste B., et al. Piezo1, a Mechanically Activated Ion Channel, Is Required for Vascular Development in Mice. Proc. Natl. Acad. Sci. USA. 2014;111:10347–10352. doi: 10.1073/pnas.1409233111. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

