Discovery of a Novel Antimicrobial Peptide from Paenibacillus sp. Na14 with Potent Activity Against Gram-Negative Bacteria and Genomic Insights into Its Biosynthetic Pathway
- PMID: 40867999
- PMCID: PMC12383022
- DOI: 10.3390/antibiotics14080805
Discovery of a Novel Antimicrobial Peptide from Paenibacillus sp. Na14 with Potent Activity Against Gram-Negative Bacteria and Genomic Insights into Its Biosynthetic Pathway
Abstract
Background/objectives: Antimicrobial resistance (AMR) contributes to millions of deaths globally each year, creating an urgent need for new therapeutic agents. Antimicrobial peptides (AMPs) have emerged as promising candidates due to their potential to combat AMR pathogens. This study aimed to evaluate the antimicrobial activity of an AMP from a soil-derived bacterial isolate against Gram-negative bacteria.
Method: Soil bacteria were isolated and screened for antimicrobial activity. The bioactive peptide was purified and determined its structure and antimicrobial efficacy. Genomic analysis was conducted to predict the biosynthetic gene clusters (BGCs) responsible for AMP production.
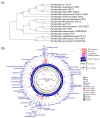
Results: Genomic analysis identified the isolate as Paenibacillus sp. Na14, which exhibited low genomic similarity (61.0%) to other known Paenibacillus species, suggesting it may represent a novel species. The AMP from the Na14 strain exhibited heat stability up to 90 °C for 3 h and retained its activity across a broad pH range from 3 to 11. Structural analysis revealed that the Na14 peptide consisted of 14 amino acid residues, adopting an α-helical structure. This peptide exhibited bactericidal activity at concentrations of 2-4 µg/mL within 6-12 h, and its killing rate was concentration-dependent. The peptide was found to disrupt the bacterial membranes. The Na14 peptide shared 64.29% sequence similarity with brevibacillin 2V, an AMP from Brevibacillus sp., which also belongs to the Paenibacillaceae family. Genomic annotation identified BGCs associated with secondary metabolism, with a particular focus on non-ribosomal peptide synthetase (NRPS) gene clusters. Structural modeling of the predicted NRPS enzymes showed high similarity to known NRPS modules in Brevibacillus species. These genomic findings provide evidence supporting the similarity between the Na14 peptide and brevibacillin 2V.
Conclusions: This study highlights the discovery of a novel AMP with potent activity against Gram-negative pathogens and provides new insight into conserved AMP biosynthetic enzymes within the Paenibacillaceae family.
Keywords: Paenibacillus sp. Na14; antimicrobial peptides; biosynthetic gene clusters; non-ribosomal peptide synthetase; soil bacteria.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analysis, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures






References
Grants and funding
LinkOut - more resources
Full Text Sources

