Umbilical Cord Mesenchymal Stem Cell-Derived Extracellular Vesicles Enhance Chondrocyte Function by Reducing Oxidative Stress in Chondrocytes
- PMID: 40869010
- PMCID: PMC12386668
- DOI: 10.3390/ijms26167683
Umbilical Cord Mesenchymal Stem Cell-Derived Extracellular Vesicles Enhance Chondrocyte Function by Reducing Oxidative Stress in Chondrocytes
Abstract
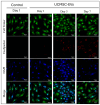
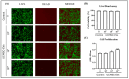
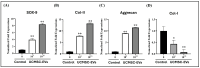
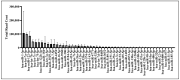
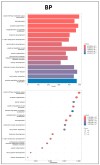
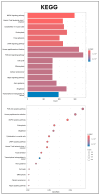
Articular cartilage (AC) has a very limited capacity for self-healing once damaged. Chondrocytes maintain AC homeostasis and are key cells in AC tissue engineering (ACTE). However, chondrocytes lose their function due to oxidative stress. Umbilical cord mesenchymal stem cells (UCMSCs) are investigated as an alternative cell source for ACTE. MSCs are known to regulate tissue regeneration through host cell modulation, largely via extracellular vesicle (EV)-mediated cell-to-cell communication. The purpose of this study was to verify whether UCMSC-derived EVs (UCMSC-EVs) enhance chondrocyte function. The mean particle sizes of the UCMSC-EVs were 79.8 ± 19.05 nm. Transmission electron microscopy (TEM) revealed that UCMSC-EVs exhibited a spherical morphology. The presence of CD9, CD63, and CD81 confirmed the identity of UCMSC-EVs, with α-tubulin undetected. UCMSC-EVs maintained chondrocyte survival, and increased chondrocyte proliferation after intake by chondrocytes. UCMSC-EVs upregulated mRNA levels of SOX-9, collagen type II (Col-II), and Aggrecan, while decreasing collagen type I (Col-I) levels. UCMSC-EVs reduced the oxidative stress of chondrocytes by reducing mitochondrial superoxide production and increasing protein levels of SOD-2 and Sirt-3 in chondrocytes. The 50 most abundant known microRNAs (miRNAs) derived from UCMSC-EVs were selected for gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses. GO analysis revealed enrichment in pathways associated with small GTPase-mediated signal transduction, GTPase regulatory activity, and mitochondrial matrix. The KEGG analysis indicated that these miRNAs may regulate chondrocyte function through the PI3K-Akt, MAPK, and cAMP signaling pathways. In summary, this study shows that UCMSC-EVs enhance chondrocyte function and may be applied to ACTE.
Keywords: articular cartilage tissue engineering (ACTE); chondrocyte function; extracellular vesicles (EVs); miRNA; oxidative stress; umbilical cord mesenchymal stem cells (UCMSCs).
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

