Microbial Signatures of Obesity-Aggravated Psoriasis: Insights from an Imiquimod-Based Mouse Model
- PMID: 40869018
- PMCID: PMC12386422
- DOI: 10.3390/ijms26167697
Microbial Signatures of Obesity-Aggravated Psoriasis: Insights from an Imiquimod-Based Mouse Model
Abstract
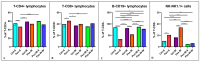
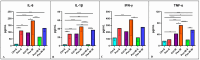
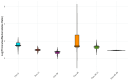
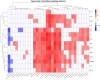
As obesity and Western diet consumption are key factors contributing to gut dysbiosis, we investigated the relationship between intestinal microbiota, obesity, and psoriasis in an imiquimod-based model. C57BL/6 mice were used as follows: psoriasis-induced groups fed continuously with a standard or Western diet, psoriasis-induced group fed with a Western diet and then returned to a standard diet, and controls. For each group, clinicopathological, immune, and metabolic parameters were integrated with microbiome data. The imiquimod-based models displayed human psoriasis features and significant changes in immune parameters. Hence, psoriatic mice on prolonged high-fat intake presented decreased microbial richness and evenness and a gut microbiome composition resembling that of obese mice. Ruminococcus, Clostridium, Desulfovibrio, and Enterorhabdus were the most abundant genera in the obesity-enhanced psoriasis group. Raoultella abundance was linked with psoriasis. Yet, the same pathobionts over-represented in the obese psoriatic mice displayed positive correlations with metabolic stress indicators and proinflammatory factors, indicating potential biomarkers of disease severity. Conversely, Lactobacillus taiwanensis, Alistipes putredinis, and Eubacterium hadrum might be potential taxa for attenuating the metabolic burden in obesity-enhanced psoriasis. Here, we depict the microbial signatures associated with inflammation and metabolic stress in an obesity-aggravated psoriasis mouse model.
Keywords: 16S rRNA sequencing; diet; dysbiosis; microbiota; psoriasis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures










References
-
- Everard A., Belzer C., Geurts L., Ouwerkerk J.P., Druart C., Bindels L.B., Guiot Y., Derrien M., Muccioli G.G., Delzenne N.M., et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA. 2013;110:9066–9071. doi: 10.1073/pnas.1219451110. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

