Transcriptomic Signatures and Molecular Pathways in Hidradenitis Suppurativa-A Narrative Review
- PMID: 40869025
- PMCID: PMC12386655
- DOI: 10.3390/ijms26167704
Transcriptomic Signatures and Molecular Pathways in Hidradenitis Suppurativa-A Narrative Review
Abstract
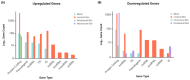
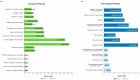
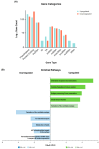
Hidradenitis suppurativa (HS) is a chronic, relapsing inflammatory dermatosis of the pilosebaceous unit characterized by nodules, abscesses, and dermal tunnels. Recent transcriptomic studies have implicated dysregulation of innate and adaptive immune responses, epidermal barrier dysfunction, and systemic metabolic alterations. This review synthesizes findings from 16 studies investigating the HS transcriptome using bulk and single-cell RNA sequencing. Differential gene expression analyses revealed extensive upregulation of inflammatory cytokines and chemokines, particularly in lesional and perilesional skin. These changes were also mirrored in non-lesional skin, suggesting diffuse immune dysregulation beyond visibly affected areas. Downregulated pathways include those involved in lipid metabolism, muscle contraction, and neuronal signaling, potentially linking HS to obesity, metabolic syndrome, and neuropsychiatric comorbidities. Single-cell transcriptomics confirmed the enrichment of keratinocytes and immune cells (B cells, plasma cells, M1 macrophages, and T cells) with proinflammatory profiles in HS lesions. Keratinocyte dysfunction further implicated a compromised epidermal barrier in disease pathogenesis. While transcriptomic studies have advanced mechanistic understanding and highlighted therapeutic targets-such as the IL-1β-TH17 axis and B cell signaling pathways-methodological heterogeneity limits cross-study comparisons. Integration of multi-omics data and standardized phenotyping will be essential to identify robust biomarkers, stratify HS subtypes, and guide personalized therapeutic approaches.
Keywords: hidradenitis suppurativa; pathomechanisms; targeted treatment; transcriptome.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Zouboulis C.C., Bechara F.G., Fritz K., Goebeler M., Hetzer F.H., Just E., Kirsten N., Kokolakis G., Kurzen H., Nikolakis G., et al. S2k guideline for the treatment of hidradenitis suppurativa/acne inversa—Short version. J. Dtsch. Dermatol. Ges. 2024;22:868–889. doi: 10.1111/ddg.15412. - DOI - PubMed
-
- Zouboulis C.C., Nogueira da Costa A., Makrantonaki E., Hou X.X., Almansouri D., Dudley J.T., Edwards H., Readhead B., Balthasar O., Jemec G.B.E., et al. Alterations in innate immunity and epithelial cell differentiation are the molecular pillars of hidradenitis suppurativa. J. Eur. Acad. Dermatol. Venereol. 2020;34:846–861. doi: 10.1111/jdv.16147. - DOI - PubMed
-
- Bouazzi D., Andersen R.K., Vinding G.R., Medianfar C.E., Nielsen S.M., Saunte D.M.L., Chandran N.S., van der Zee H.H., Zouboulis C.C., Benhadou F., et al. The Global Hidradenitis Suppurativa Atlas Methodology: Combining Global Proportions in a Pooled Analysis. Dermatology. 2024;240:369–375. doi: 10.1159/000536389. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

