Construction of a Genetic Prognostic Model in the Glioblastoma Tumor Microenvironment
- PMID: 40869909
- PMCID: PMC12385252
- DOI: 10.3390/genes16080861
Construction of a Genetic Prognostic Model in the Glioblastoma Tumor Microenvironment
Abstract
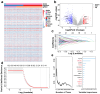
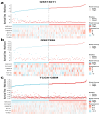
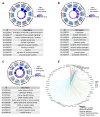
Background: Glioblastoma (GBM) is one of the most challenging malignancies in all of neoplasms. These malignancies are associated with unfavorable clinical outcomes and significantly compromised patient wellbeing. The immunological landscape within the tumor microenvironment (TME) plays a critical role in determining GBM prognosis. By mining data from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases and correlating them with immune responses in the TME, genes associated with the immune microenvironment with potential prognostic value were obtained. Method: We selected GSE16011 as the training set. Gene expression profiles were substrates scored by both ESTIMATE and xCell, and immune cell subpopulations in GBM were analyzed by CIBERSORT. Gene expression profiles associated with low immune scores were performed by lasso regression, Cox analysis and random forest (RF) to identify a prognostic model for the multiple genes associated with immune infiltration in GBM. Then we constructed a nomogram to optimize the prognostic model using GSE7696 and TCGA-GBM as validation sets and evaluated these data for gene mutation and gene enrichment analysis.
Result: The prognostic correlation between the six genes (MEOX2, PHYHIP, RBBP8, ST18, TCF12, and THRB) and GBM was finally found by lasso regression, Cox regression, and RF, and the online database obtained that all six genes were differentially expressed in GBM. Therefore, a prognostic correlation model was constructed based on the six genes. Kaplan-Meier (KM) survival analysis showed that this prognostic model had excellent prognostic ability.
Conclusions: Prognostic models based on tumor microenvironment and immune score stratification and the construction of related genes have potential applications for prognostic analysis of GBM patients.
Keywords: glioblastoma; nomogram; prognosis; tumor microenvironment.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures







References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

