Local Fungi Promote Plant Growth by Positively Affecting Rhizosphere Metabolites to Drive Beneficial Microbial Assembly
- PMID: 40871256
- PMCID: PMC12388802
- DOI: 10.3390/microorganisms13081752
Local Fungi Promote Plant Growth by Positively Affecting Rhizosphere Metabolites to Drive Beneficial Microbial Assembly
Abstract
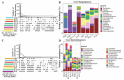
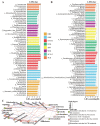
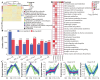
Ecological restoration in the cold and high-altitude mining areas of the Qinghai-Tibet Plateau is faced with dual challenges of extreme environments and insufficient microbial adaptability. This study aimed to screen local microbial resources with both extreme environmental adaptability and plant-growth-promoting functions. Local fungi (DK; F18-3) and commercially available bacteria (B0) were used as materials to explore their regulatory mechanisms for plant growth, soil physicochemical factors, microbial communities, and metabolic profiles in the field. Compared to bacterial treatments, local fungi treatments exhibited stronger ecological restoration efficacy. In addition, the DK and F18-3 strains, respectively, increased shoot and root biomass by 23.43% and 195.58% and significantly enhanced soil nutrient content and enzyme activity. Microbiome analysis further implied that, compared with the CK, DK treatment could significantly improve the α-diversity of fungi in the rhizosphere soil (the Shannon index increased by 14.27%) and increased the amount of unique bacterial genera in the rhizosphere soil of plants, totaling fourteen genera. Meanwhile, this aggregated the most biomarkers and beneficial microorganisms and strengthened the interactions among beneficial microorganisms. After DK treatment, twenty of the positively accumulated differential metabolites (DMs) in the plant rhizosphere were highly positively associated with six plant traits such as shoot length and root length, as well as beneficial microorganisms (e.g., Apodus and Pseudogymnoascus), but two DMs were highly negatively related to plant pathogenic fungi (including Cistella and Alternaria). Specifically, DK mainly inhibited the growth of pathogenic fungi through regulating the accumulation of D-(+)-Malic acid and Gamma-Aminobutyric acid (Cistella and Alternaria decreased by 84.20% and 58.53%, respectively). In contrast, the F18-3 strain mainly exerted its antibacterial effect by enriching Acidovorax genus microorganisms. This study verified the core role of local fungi in the restoration of mining areas in the Qinghai-Tibet Plateau and provided a new direction for the development of microbial agents for ecological restoration in the Qinghai-Tibet Plateau.
Keywords: beneficial microorganisms; ecological restoration; local fungi; promoting plant growth; rhizosphere metabolites.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Feng Y., Wang J., Bai Z., Reading L. Effects of surface coal mining and land reclamation on soil properties: A review. Earth-Sci. Rev. 2019;191:12–25. doi: 10.1016/j.earscirev.2019.02.015. - DOI
-
- Ge H., Feng Y., Li Y., Yang W.L., Gong N. Heavy metal pollution diagnosis and ecological risk assessment of the surrounding soils of coal waste pile at Naluo Coal Mine, Liupanshui, Guizhou. Int. J. Min. Reclam. Environ. 2016;30:312–318. doi: 10.1080/17480930.2015.1050840. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

