Characterization of a Thermostable α-Amylase from Bacillus licheniformis 104.K for Industrial Applications
- PMID: 40871262
- PMCID: PMC12388735
- DOI: 10.3390/microorganisms13081757
Characterization of a Thermostable α-Amylase from Bacillus licheniformis 104.K for Industrial Applications
Abstract
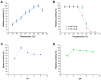
This study describes the characterization of a novel thermostable α-amylase from a Bacillus licheniformis 104.K strain isolated from the Kashkadarya region of Uzbekistan. Phylogenetic analysis revealed that the thermostable α-amylase belongs to glycoside hydrolase family 13 subfamily 5 (GH13_5) and shares high sequence similarity with known α-amylases. Our results demonstrate that the recombinant α-amylase exhibits optimal activity at pH 6.0 and 90 °C, retaining full activity after 30 min at 60 °C. The addition of CaCl2 significantly enhanced thermostability, with the enzyme retaining more than 95% of its initial activity at 70 °C after 30 min. Our findings indicate that α-amylase from B. licheniformis 104.K is a functional, thermostable enzyme with potential industrial applications. This study highlights the commercial significance of thermostable amylases and the need to identify novel, cost-effective, and sustainable sources. The results of this study will contribute to the fields of enzyme applications, stabilizing additives, and genetic engineering of thermostable genes.
Keywords: Bacillus licheniformis; GH13_5 subfamily; affinity chromatography; recombinant enzyme; thermostability; α-amylase.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Green starch nanoparticles production in situ using α-amylase from a newly isolated Bacillus subtilis strain-MA6: statistical designs and characterizations.Microb Cell Fact. 2025 Aug 18;24(1):187. doi: 10.1186/s12934-025-02812-y. Microb Cell Fact. 2025. PMID: 40826077 Free PMC article.
-
Biochemical and thermodynamic characterization of a novel α-amylase from Avena fatua for biotechnological applications.Bioorg Chem. 2025 Aug 18;164:108883. doi: 10.1016/j.bioorg.2025.108883. Online ahead of print. Bioorg Chem. 2025. PMID: 40845552
-
Isolation, expression, and in silico profiling of a thermostable xylanase from Geobacillus stearothermophilus strain NASA267: insights into structural features and agro-waste valorization.Microb Cell Fact. 2025 Mar 21;24(1):69. doi: 10.1186/s12934-025-02672-6. Microb Cell Fact. 2025. Retraction in: Microb Cell Fact. 2025 Jul 10;24(1):161. doi: 10.1186/s12934-025-02790-1. PMID: 40119336 Free PMC article. Retracted.
-
Hunt for α-amylase from metagenome and strategies to improve its thermostability: a systematic review.World J Microbiol Biotechnol. 2022 Aug 24;38(11):203. doi: 10.1007/s11274-022-03396-0. World J Microbiol Biotechnol. 2022. PMID: 35999473
-
Adefovir dipivoxil and pegylated interferon alfa-2a for the treatment of chronic hepatitis B: a systematic review and economic evaluation.Health Technol Assess. 2006 Aug;10(28):iii-iv, xi-xiv, 1-183. doi: 10.3310/hta10280. Health Technol Assess. 2006. PMID: 16904047
References
Grants and funding
- IL-4821091620/Agency of Innovative Development of the Republic of Uzbekistan
- 2022-1.2.2-TÉT-IPARI-UZ-2022-00003/National Research, Development and Innovation Office of Hungary
- Bolyai János Research Scholarship to GNN/Hungarian Academy of Sciences
- Somogyi Mihály Grant to GNN/Varga József Foundation, Budapest University of Technology and Economics
LinkOut - more resources
Full Text Sources

