Degradation of Synthetic and Natural Textile Materials Using Streptomyces Strains: Model Compost and Genome Exploration for Potential Plastic-Degrading Enzymes
- PMID: 40871304
- PMCID: PMC12388512
- DOI: 10.3390/microorganisms13081800
Degradation of Synthetic and Natural Textile Materials Using Streptomyces Strains: Model Compost and Genome Exploration for Potential Plastic-Degrading Enzymes
Abstract
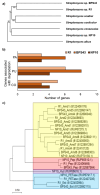
Given the environmental significance of the textile industry, especially the accumulation of nondegradable materials, there is extensive development of greener approaches to fabric waste management. Here, we investigated the biodegradation potential of three Streptomyces strains in model compost on polyamide (PA) and polyamide-elastane (PA-EA) as synthetic, and on cotton (CO) as natural textile materials. Weight change of the materials was followed, while Fourier-Transform Infrared Spectroscopy (FTIR) and Scanning Electron Microscopy (SEM) were used to analyze surface changes of the materials upon biodegradation. The bioluminescence-based toxicity test employing Aliivibrio fischeri confirmed the ecological safety of the tested textiles. After 12 months, the increase of 10 and 16% weight loss, of PA-EA and PA, respectively, was observed in compost augmented with Streptomyces sp. BPS43. Additionally, a 14% increase in cotton degradation was recorded after 2 months in compost augmented with Streptomyces sp. NP10. Genome exploration of the strains was carried out for potential plastic-degrading enzymes. It highlighted BPS43 as the most versatile strain with specific amidases that show sequence identity to UMG-SP-1, UMG-SP-2, and UMG-SP-3 (polyurethane degrading enzymes identified from compost metagenome). Our results showcase the behavior of Streptomyces sp. BPS43 in the degradation of PA and PA-EA textiles in composting conditions, with enzymatic potential that could be further characterized and optimized for increased synthetic textile degradation.
Keywords: Streptomyces; bioremediation; compost; degradation; genome analysis; polyamide; textile.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





Similar articles
-
Carboxylic ester hydrolases from Antarctic psychrophilic Psychrobacter strains: From genome prospecting to biotreatment of polyester plastics.Bioresour Technol. 2025 Nov;436:133052. doi: 10.1016/j.biortech.2025.133052. Epub 2025 Aug 5. Bioresour Technol. 2025. PMID: 40752649
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Does Augmenting Irradiated Autografts With Free Vascularized Fibula Graft in Patients With Bone Loss From a Malignant Tumor Achieve Union, Function, and Complication Rate Comparably to Patients Without Bone Loss and Augmentation When Reconstructing Intercalary Resections in the Lower Extremity?Clin Orthop Relat Res. 2025 Jun 26;483(9):1680-1695. doi: 10.1097/CORR.0000000000003599. Clin Orthop Relat Res. 2025. PMID: 40569278
-
Antidepressants for pain management in adults with chronic pain: a network meta-analysis.Health Technol Assess. 2024 Oct;28(62):1-155. doi: 10.3310/MKRT2948. Health Technol Assess. 2024. PMID: 39367772 Free PMC article.
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
References
-
- Foundation C.E. The Circularity Gap Report. [(accessed on 24 January 2024)]. Available online: https://www.circularity-gap.world/2024.
-
- Exchange T. 11th Edition of Textile Exchange’s Annual Materials Market Report. [(accessed on 26 September 2024)]. Available online: https://textileexchange.org/app/uploads/2024/09/Materials-Market-Report-....
-
- Roundup T. 17 Most Worrysing Textile Waste Statistics & Facts. [(accessed on 18 March 2024)]. Available online: https://theroundup.org/textile-waste-statistics/
-
- Yadav P., Mishra V. Comprehending microplastic pollution in diverse environment: Assessing fate, impacts, and remediation approaches. Int. Biodeterior. Biodegrad. 2025;196:105953. doi: 10.1016/j.ibiod.2024.105953. - DOI
-
- UNEP Sustainable Fashion to Take Centre Stage on Zero Waste Day. [(accessed on 7 February 2025)]. Available online: https://www.unep.org/technical-highlight/sustainable-fashion-take-centre....
Grants and funding
LinkOut - more resources
Full Text Sources

