Environmental Factors Drive the Changes of Bacterial Structure and Functional Diversity in Rhizosphere Soil of Hippophae rhamnoides subsp. sinensis Rousi in Arid Regions of Northwest China
- PMID: 40871364
- PMCID: PMC12388428
- DOI: 10.3390/microorganisms13081860
Environmental Factors Drive the Changes of Bacterial Structure and Functional Diversity in Rhizosphere Soil of Hippophae rhamnoides subsp. sinensis Rousi in Arid Regions of Northwest China
Abstract
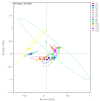
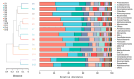
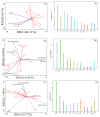
Hippophae rhamnoides subsp. sinensis Rousi has high ecological and medicinal value, and it is an important plant resource unique to the arid regions of Northwest China. Exploring the influence of climate characteristics and soil factors on the composition, diversity, and function of the rhizosphere bacterial community of Chinese seabuckthorn is of great value for developing and popularizing characteristic plant resources in the arid regions of Northwest China. In this study, the rhizosphere soil of 13 Chinese seabuckthorn distribution areas in the northwest of China was taken as the research object, the bacterial community map was constructed based on 16S rRNA gene high-throughput sequencing technology, and the species abundance composition, structural diversity, molecular co-occurrence network, and phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt), as well as the function of rhizosphere soil bacterial community, were systematically studied. Combined with Mantel test and redundancy analysis (RDA), the key habitat factors driving the rhizosphere soil bacterial community structure of Chinese seabuckthorn were explored. The results showed that: (1) The number of amplicon sequence variants (ASVs) in rhizosphere soil bacterial community of Chinese seabuckthorn was the highest in S2(3072) and the S12(3637), and the lowest in the S11(1358) and S13(1996). The rhizosphere soil bacterial community was primarily composed of Proteobacteria, Actinobacteriota, and Acidobacteriota. Except for the S6 and S11 habitats, the dominant bacterial genera were mainly Achromobacter, Acidobacter (RB41), and Sphingomonas. (2) The α and β diversity of rhizosphere soil bacterial communities of Chinese seabuckthorn across 13 distribution areas were significantly different. The number of operational taxonomic units (OTUs), Ace index, and Chao 1 index of soil bacterial community in the S12 distribution area are the highest, and they are the lowest in S11 distribution area, with significant differences. The aggregation of soil bacterial communities in the S5 and S10 distribution areas is the highest, while it is the lowest in the S6 and S11 distribution areas. (3) PICRUSt function classification of soil bacteria showed that Metabolism and Genetic Information Processing functions were the strongest across all distribution areas, with S10 exhibiting higher functional capacity than other areas and S11 showing the weakest. (4) Cluster analysis revealed that soil bacteria across the 13 distribution areas were clustered into two groups, with S10 and S12 distribution areas as one group (Group 1) and the remaining 11 distribution areas as another group (Group 2). (5) Redundancy analysis revealed that pH was the key soil environmental factor driving the rhizosphere soil bacterial community α-diversity of Chinese seabuckthorn, followed by altitude (ALT) and soil water content (SWC). In summary, Chinese seabuckthorn prefers neutral to alkaline soils, and environmental factors play an important role in driving bacterial diversity, community structure, functional profiles, and co-occurrence networks in rhizosphere soil of Chinese seabuckthorn.
Keywords: Hippophae rhamnoides subsp. sinensis Rousi; function prediction; geographical pattern; pH; rhizosphere soil.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures










References
-
- Li X.-w., Sun K., Li Y.-h. Variation in leaf nitrogen and phosphorus stoichiometry in the nitrogen-fixing Chinese sea-buckthorn (Hippophae rhamnoides L. subsp. sinensis Rousi) across northern China. Ecol. Res. 2014;29:723–731. doi: 10.1007/s11284-014-1165-y. - DOI
-
- Cheng Z.-y., Chen Y.-f., Yuan H.-j. Molecular mechanisms of seed germination in Hippophae rhamnoides L. based on transcriptomics. Res. Cold Arid. Reg. 2024;16:310–322. doi: 10.1016/j.rcar.2024.11.003. - DOI
-
- Su Y., Li S.-x., Jiang H., Duan B.-l., Liu M.-y., Zhang Y.-b. Sex-specific physiological and growth responses to elevated temperature and CO2 concentration in Chinese seabuckthorn (Hippophae rhamnoides subsp. sinensis Rousi) Acta Physiol. Plant. 2023;45:53. doi: 10.1007/s11738-023-03520-z. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

