Why Are Long-Read Sequencing Methods Revolutionizing Microbiome Analysis?
- PMID: 40871366
- PMCID: PMC12388134
- DOI: 10.3390/microorganisms13081861
Why Are Long-Read Sequencing Methods Revolutionizing Microbiome Analysis?
Abstract
Most of the knowledge available on the composition and functionality of microbial communities in different ecosystems comes from short-read sequencing methods. It implies limitations regarding taxonomic resolution, variant detection, and genome assembly contiguity. Long-read sequencing technologies can overcome these limitations, transforming the analysis of microbial community composition and functionality. It is essential to understand the characteristics of each sequencing technology to select the most suitable one for each microbiome study. This review aims to show how long-read sequencing methods have revolutionized microbiome analysis in ecosystems and to provide a practical tool for selecting sequencing methods. To this end, the evolution of sequencing technologies, their advantages and disadvantages for microbiome studies, and the new dimensions enabled by long-read sequencing technologies, such as virome and epigenetic analysis, are described. Moreover, desirable characteristics for microbiome sequencing technologies are proposed, including a visual comparison of available sequencing platforms. Finally, amplicon and metagenomics approaches and the sequencing depth are discussed when using long-read sequencing technologies in microbiome studies. In conclusion, although no single sequencing method currently possesses all the ideal features for microbiome analysis in ecosystems, long-read sequencing technologies represent an advancement in key aspects, including longer read lengths, higher accuracy, shorter runtimes, higher output, more affordable costs, and greater portability. Therefore, more research using long-read sequencing is recommended to strengthen its application in microbiome analysis.
Keywords: 16S; ecosystems; genome assembly; long-read sequencing; metagenomics; microbiome; taxonomic resolution; variants.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
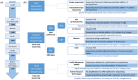

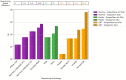


Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Healthcare workers' informal uses of mobile phones and other mobile devices to support their work: a qualitative evidence synthesis.Cochrane Database Syst Rev. 2024 Aug 27;8(8):CD015705. doi: 10.1002/14651858.CD015705.pub2. Cochrane Database Syst Rev. 2024. PMID: 39189465 Free PMC article.
-
De novo Genome Assembly Using Long Reads and Chromosome Conformation Capture.Methods Mol Biol. 2025;2935:1-27. doi: 10.1007/978-1-0716-4583-3_1. Methods Mol Biol. 2025. PMID: 40828272
References
-
- Klinsawat W., Uthaipaisanwong P., Jenjaroenpun P., Sripiboon S., Wongsurawat T., Kusonmano K. Microbiome Variations among Age Classes and Diets of Captive Asian Elephants (Elephas maximus) in Thailand Using Full-Length 16S rRNA Nanopore Sequencing. Sci. Rep. 2023;13:17685. doi: 10.1038/s41598-023-44981-z. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

