Microbial Assembly and Stress-Tolerance Mechanisms in Salt-Adapted Plants Along the Shore of a Salt Lake: Implications for Saline-Alkaline Soil Remediation
- PMID: 40871446
- PMCID: PMC12388437
- DOI: 10.3390/microorganisms13081942
Microbial Assembly and Stress-Tolerance Mechanisms in Salt-Adapted Plants Along the Shore of a Salt Lake: Implications for Saline-Alkaline Soil Remediation
Abstract
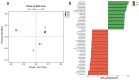
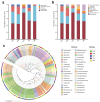
Investigating the microbial community structure and stress-tolerance mechanisms in the rhizospheres of salt-adapted plants along saline lakes is critical for understanding plant-microbe interactions in extreme environments and developing effective strategies for saline-alkaline soil remediation. This study explored the rhizosphere microbiomes of four salt-adapted species (Suaeda glauca, Artemisia carvifolia, Chloris virgata, and Limonium bicolor) from the Yuncheng Salt Lake region in China using high-throughput sequencing. Cultivable salt-tolerant plant growth-promoting rhizobacteria (PGPR) were isolated and characterized to identify functional genes related to stress resistance. Results revealed that plant identity and soil physicochemical properties jointly shaped the microbial community composition, with total organic carbon being a dominant driver explaining 17.6% of the variation. Cyanobacteria dominated low-salinity environments, while Firmicutes thrived in high-salinity niches. Isolated PGPR strains exhibited tolerance up to 15% salinity and harbored genes associated with heat (htpX), osmotic stress (otsA), oxidative stress (katE), and UV radiation (uvrA). Notably, Peribacillus and Isoptericola strains demonstrated broad functional versatility and robust halotolerance. Our findings highlight that TOC (total organic carbon) plays a pivotal role in microbial assembly under extreme salinity, surpassing host genetic influences. The identified PGPR strains, with their stress-resistance traits and functional gene repertoires, hold significant promise for biotechnological applications in saline-alkaline soil remediation and sustainable agriculture.
Keywords: Yuncheng salt lake; plant growth-promoting rhizobacteria (PGPR); saline-alkaline soil remediation; salt tolerance mechanisms; salt-adapted plant rhizosphere microbiome; total organic carbon.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Wang J., Yasen M., Gong M., Zhou Q., Li M. Structural variability in the rhizosphere bacterial communities of three halophytes under different levels of salinity-alkalinity. Plant Soil. 2024;502:709–723. doi: 10.1007/s11104-024-06580-3. - DOI
-
- Xavier J.F., da Costa D.P., da Silva Gonçalves J.V., Pinto M.C.F., Goulart R.S., Zonta E., da Silva Coelho I. Different halophytes orchestrate microbial diversity in the rhizosphere of salinity-impacted soils. Appl. Soil. Ecol. 2024;202:105588. doi: 10.1016/j.apsoil.2024.105588. - DOI
-
- Garbeva P., van Elsas J.D., van Veen J.A. Rhizosphere microbial community and its response to plant species and soil history. Plant Soil. 2008;302:19–32. doi: 10.1007/s11104-007-9432-0. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

