Outbreak Caused by VIM-1- and VIM-4-Positive Proteus mirabilis in a Hospital in Zagreb
- PMID: 40872247
- PMCID: PMC12389321
- DOI: 10.3390/pathogens14080737
Outbreak Caused by VIM-1- and VIM-4-Positive Proteus mirabilis in a Hospital in Zagreb
Abstract
Background/objectives: Proteus mirabilis is a frequent causative agent of urinary and wound infections in both community and hospital settings. It develops resistance to expanded-spectrum cephalosporins (ESCs) due to the production of extended-spectrum β-lactamases (ESBLs) or plasmid-mediated AmpC β-lactamases (p-AmpCs). Recently, carbapenem-resistant isolates of P. mirabilis emerged due to the production of carbapenemases, mostly belonging to Ambler classes B and D. Here, we report an outbreak of infections due to carbapenem-resistant P. mirabilis that were observed in a psychiatric hospital in Zagreb, Croatia. The characteristics of ESBL and carbapenemase-producing P. mirabilis isolates, associated with an outbreak, were analyzed.
Materials and methods: The antibiotic susceptibility testing was performed by the disk-diffusion and broth dilution methods. The double-disk synergy test (DDST) and inhibitor-based test with clavulanic and phenylboronic acid were applied to screen for ESBLs and p-AmpCs, respectively. Carbapenemases were screened by the modified Hodge test (MHT), while carbapenem hydrolysis was investigated by the carbapenem inactivation method (CIM) and EDTA-carbapenem-inactivation method (eCIM). The nature of the ESBLs, carbapenemases, and fluoroquinolone-resistance determinants was investigated by PCR. Plasmids were characterized by PCR-based replicon typing (PBRT). Selected isolates were subjected to molecular characterization of the resistome by an Inter-Array Genotyping Kit CarbaResisit and whole-genome sequencing (WGS).
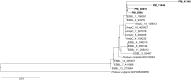
Results: In total, 20 isolates were collected and analyzed. All isolates exhibited resistance to amoxicillin alone and when combined with clavulanic acid, cefuroxime, cefotaxime, ceftriaxone, cefepime, imipenem, ceftazidime-avibactam, ceftolozane-tazobactam, gentamicin, amikacin, and ciprofloxacin. There was uniform susceptibility to ertapenem, meropenem, and cefiderocol. The DDST and combined disk test with clavulanic acid were positive, indicating the production of an ESBL. The MHT was negative in all except one isolate, while the CIM showed moderate sensitivity, but only with imipenem as the indicator disk. Furthermore, eCIM tested positive in all of the CIM-positive isolates, consistent with a metallo-β-lactamase (MBL). PCR and sequencing of the selected amplicons identified VIM-1 and VIM-4. The Inter-Array Genotyping Kit CarbaResist and WGS identified β-lactam resistance genes blaVIM, blaCTX-M-15, and blaTEM genes; aminoglycoside resistance genes aac(3)-IId, aph(6)-Id, aph(3″)-Ib, aadA1, armA, and aac(6')-IIc; as well as resistance genes for sulphonamides sul1 and sul2, trimethoprim dfr1, chloramphenicol cat, and tetracycline tet(J).
Conclusions: This study revealed an epidemic spread of carbapenemase-producing P. mirabilis in two wards in a psychiatric hospital. Due to the extensively resistant phenotype (XDR), therapeutic options were limited. This is the first report of carbapenemase-producing P. mirabilis in Croatia.
Keywords: CTX-M-15; Proteus mirabilis; VIM; epidemic spread; multidrug resistance.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Sanches M.S., Silva L.C., Silva C.R.D., Montini V.H., Oliva B.H.D., Guidone G.H.M., Nogueira M.C.L., Menck-Costa M.F., Kobayashi R.K.T., Vespero E.C., et al. Prevalence of Antimicrobial Resistance and Clonal Relationship in ESBL/AmpC-Producing Proteus mirabilis Isolated from Meat Products and Community-Acquired Urinary Tract Infection (UTI-CA) in Southern Brazil. Antibiotics. 2023;10:370. doi: 10.3390/antibiotics12020370. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous