Virome Survey of Banana Plantations and Surrounding Plants in Malawi
- PMID: 40872785
- PMCID: PMC12390665
- DOI: 10.3390/v17081068
Virome Survey of Banana Plantations and Surrounding Plants in Malawi
Abstract
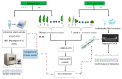
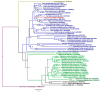
A virome survey of banana plantations and their surrounding plants was carried out at nation-wide level in Malawi using virion associated nucleic acids (VANA) high throughput sequencing (HTS) on pooled samples and appropriate alien controls. In total, 366 plants were sequenced, and 23 plant virus species were detected, three species on banana (275 plants) and 20 species in surrounding plants (91 plants). Two putative novel virus species; ginger tymo-like virus and pepper derived totivirus were detected and confirmed by RT-PCR on ginger and pepper. Nine known virus species and detected a host plant was identified for two of them. No viral exchange between banana and surrounding plants was observed. Results from the VANA protocol, applied to pooled banana samples, were compared with previous targeted PCR results obtained from individual banana samples. HTS test detected better BanMMV than IC-(RT)-PCR on individual samples (better inclusivity) but detected with much lower sensitivity BBTV and BSV species, often with less than 10 reads per sample. Detection of novel and known viruses and new host plants calls for strengthened sanitory and phytosanitory measures within and beyond banana production systems. Our research confirms that HTS sensitivity depends on sampling, pooling protocol and targeted virus species.
Keywords: detection; high throughput sequencing; new host plant; novel viruses; plant viruses; virome survey.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. Furthermore, we declare that no animals were used in this research. All authors have consented to the submission of this paper.
Figures





References
-
- Soko M.M. Ph.D. Thesis. Queensland University of Technology; Brisbane, Australia: 2022. Evaluation of Transgenic RNAi Banana and Plantain Lines for Resistance to Banana Bunchy Top Disease.
-
- Omondi B.A., Soko M.M., Nduwimana I., Delano R.T., Niyongere C., Simbare A., Kachigamba D., Staver C. The effectiveness of consistent roguing in managing banana bunchy top disease in smallholder production in Africa. Plant Pathol. 2020;69:1754–1766. doi: 10.1111/ppa.13253. - DOI
-
- Masangwa J.I.G., Pareta N.F., Moses P., Hřibová E., Doležel J., Fandika I., Massart S. Surveillance and Molecular Characterization of Banana Viruses and Their Association with Musa Germplasm in Malawi. bioRxiv. 2024 doi: 10.1101/2024.06.24.600487. - DOI
-
- Ndunguru J., Sseruwagi P., Tairo F., Stomeo F., Maina S., Djinkeng A., Kehoe M., Boykin L.M., Melcher U. Analyses of Twelve New Whole Genome Sequences of Cassava Brown Streak Viruses and Ugandan Cassava Brown Streak Viruses from East Africa: Diversity, Supercomputing and Evidence for Further Speciation. PLoS ONE. 2015;10:e0139321. doi: 10.1371/journal.pone.0139321. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

