Identification and Structural Characterization of Viroporins from Deadly Hemorrhagic Viruses
- PMID: 40872833
- PMCID: PMC12390637
- DOI: 10.3390/v17081120
Identification and Structural Characterization of Viroporins from Deadly Hemorrhagic Viruses
Abstract
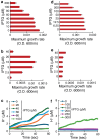
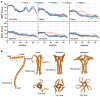
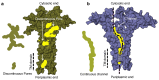
Crimean-Congo hemorrhagic fever virus (CCHF-V) and Ebola virus are lethal pathogens that cause widespread outbreaks of hemorrhagic fever. Both diseases can be transmitted through contact with the bodily fluids of infected individuals, but as an arbovirus, CCHF-V is primarily transmitted through tick bites. Both of these viruses are classified as Risk Group 4 due to the appreciable health threat they pose. To date, there are few effective treatments available to combat these deadly hemorrhagic fevers. Consequently, identifying and characterizing ion channels (viroporins) encoded in the viral genomes may lead to potential targeted drug development. Therefore, using bacteria-based genetic assays, two viroporin candidates from CCHF-V and Ebola have been examined, and their proposed structures have been modeled to aid in further drug discovery. The results indicate that CCHF-V-gp exhibits channel activity, which is indistinguishable from established viroporins found in other viruses. In contrast, our experimental approach was unable to uncover a viroporin candidate in the Ebola virus.
Keywords: Crimean–Congo hemorrhagic fever; Ebola virus disease; ion channels; structural analysis; viroporins.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Fields B.N. Fields’ Virology. Volume 1 Lippincott Williams & Wilkins; Philadelphia, PA, USA: 2007.
-
- Ergonul O., Whitehouse C.A. Crimean-Congo Hemorrhagic Fever: A Global Perspective. Springer Science & Business Media; Berlin/Heidelberg, Germany: 2007.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

