Integrated Membrane Yeast Two-Hybrid System for the Analysis of Membrane Protein Complexes
- PMID: 40873472
- PMCID: PMC12378420
- DOI: 10.21769/BioProtoc.5418
Integrated Membrane Yeast Two-Hybrid System for the Analysis of Membrane Protein Complexes
Abstract
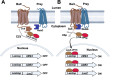
Protein-protein interactions facilitate cellular functions through the creation of networks and multi-protein complexes. Mapping the interactions within and between protein networks and elucidating the composition of protein complexes provides critical insight into biological processes. Interactions among soluble cytoplasmic proteins have been extensively investigated through the application of immunoaffinity capture as well as conventional nuclear two-hybrid testing. The integrated membrane yeast two-hybrid provides a method to investigate protein-protein interactions between integral membrane proteins in their native membrane environment. This procedure makes use of the ability of the amino-terminal fragment of ubiquitin (Nub) and the carboxyl-terminal fragment of ubiquitin (Cub) to refold reconstituting functional ubiquitin, which can be recognized by a ubiquitin peptidase. Appending a fusion protein composed of Cub fused to LexA and VP16 (CLV) to a candidate "bait" protein and Nub to candidate "prey" proteins allows a test of their interaction. If the two proteins interact closely, the CLV fragment is cleaved and enters the nucleus to activate the expression of reporter genes, signaling the interaction. When the bait and prey proteins are tagged with CLV and NubG, respectively, at their genomic loci, they are only copies of the bait and prey in the cell and are expressed under the regulation of their native promoters. This avoids overexpression artifacts that can occur if the tagged proteins are expressed from plasmids while the untagged chromosomally encoded copies of the bait and prey continue to be expressed. Key features • Allows an in vivo interaction test with integral membrane proteins in the native membrane environment. • Allows integration of NubG tag at the amino or carboxyl-terminus of prey proteins. • Avoids overexpression artifacts that can be caused by expression of CLV-tagged bait and NubG-tagged prey proteins from plasmid-based systems. • Avoids competition from untagged chromosomally encoded bait and prey proteins, as occurs when CLV-tagged bait and NubG-tagged prey are expressed from plasmids.
Keywords: MYTH; Membrane protein; Protein-protein interaction; Split-ubiquitin; Two-hybrid; Yeast; iMYTH.
©Copyright : © 2025 The Authors; This is an open access article under the CC BY-NC license.
Conflict of interest statement
Competing interestsThe authors declare no conflicts of interest.
Figures



References
-
- Wang S., Wu R., Lu J., Jiang Y., Huang T. and Cai Y.(2022). Protein‐protein interaction networks as miners of biological discovery. Proteomics. 22: e202100190. https://doi.org/ 10.1002/pmic.202100190 - DOI - PubMed
LinkOut - more resources
Full Text Sources

