MedAlmighty: enhancing disease diagnosis with large vision model distillation
- PMID: 40873493
- PMCID: PMC12378157
- DOI: 10.3389/frai.2025.1527980
MedAlmighty: enhancing disease diagnosis with large vision model distillation
Abstract
Introduction: Accurate disease diagnosis is critical in the medical field, yet it remains a challenging task due to the limited, heterogeneous, and complex nature of medical data. These challenges are particularly pronounced in multimodal tasks requiring the integration of diverse data sources. While lightweight models offer computational efficiency, they often lack the comprehensive understanding necessary for reliable clinical predictions. Conversely, large vision models, trained on extensive general-domain datasets, provide strong generalization but fall short in specialized medical applications due to domain mismatch and limited medical data availability.
Methods: To bridge the gap between general and specialized performance, we propose MedAlmighty, a knowledge distillation-based framework that synergizes the strengths of both large and small models. In this approach, we utilize DINOv2-a pre-trained large vision model-as a frozen teacher, and a lightweight convolutional neural network (CNN) as the trainable student. The student model is trained using both hard labels from the ground truth and soft targets generated by the teacher model. We adopt a hybrid loss function that combines cross-entropy loss (for classification accuracy) and Kullback-Leibler divergence (for distillation), enabling the student model to capture rich semantic features while remaining efficient and domain-aware.
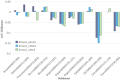
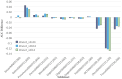
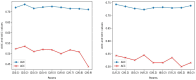
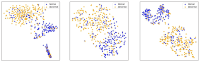
Results: Experimental evaluations reveal that MedAlmighty significantly improves disease diagnosis performance across datasets characterized by sparse and diverse medical data. The proposed model outperforms baselines by effectively integrating the generalizable representations of large models with the specialized knowledge from smaller models. The results confirm improved robustness and accuracy in complex diagnostic scenarios.
Discussion: The MedAlmighty framework demonstrates that incorporating general-domain representations via frozen large vision models-when guided by task-specific distillation strategies-can enhance the performance of lightweight medical models. This approach offers a promising solution to data scarcity and domain gap issues in medical imaging. Future work may explore extending this distillation strategy to other medical modalities and incorporating multimodal alignment for even richer representation learning.
Keywords: disease diagnosis; domain generalization; knowledge distillation; large vision model; model capacity.
Copyright © 2025 Ren, Gu and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Distilling knowledge from graph neural networks trained on cell graphs to non-neural student models.Sci Rep. 2025 Aug 10;15(1):29274. doi: 10.1038/s41598-025-13697-7. Sci Rep. 2025. PMID: 40784939 Free PMC article.
-
Enhancing Clinical Relevance of Pretrained Language Models Through Integration of External Knowledge: Case Study on Cardiovascular Diagnosis From Electronic Health Records.JMIR AI. 2024 Aug 6;3:e56932. doi: 10.2196/56932. JMIR AI. 2024. PMID: 39106099 Free PMC article.
-
Artificial intelligence for diagnosing exudative age-related macular degeneration.Cochrane Database Syst Rev. 2024 Oct 17;10(10):CD015522. doi: 10.1002/14651858.CD015522.pub2. Cochrane Database Syst Rev. 2024. PMID: 39417312
-
Magnetic resonance perfusion for differentiating low-grade from high-grade gliomas at first presentation.Cochrane Database Syst Rev. 2018 Jan 22;1(1):CD011551. doi: 10.1002/14651858.CD011551.pub2. Cochrane Database Syst Rev. 2018. PMID: 29357120 Free PMC article.
References
-
- Arumugam M., Thiyagarajan A., Adhi L., Alagar S. (2024). Crossover smell agent optimized multilayer perceptron for precise brain tumor classification on mri images. Expert Syst. Appl. 238:121453. 10.1016/j.eswa.2023.121453 - DOI
-
- Bao H., Dong L., Piao S., Wei F. (2021). Beit: bert pre-training of image transformers. arXiv preprint arXiv:2106.08254.
-
- Caron M., Touvron H., Misra I., Jégou H., Mairal J., Bojanowski P., et al. (2021). “Emerging properties in self-supervised vision transformers,” in Proceedings of the IEEE/CVF International Conference on Computer Vision, 9650–9660. 10.1109/ICCV48922.2021.00951 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

