Quantitative metagenomics for marine prokaryotes and photosynthetic eukaryotes
- PMID: 40873785
- PMCID: PMC12378644
- DOI: 10.1093/ismeco/ycaf131
Quantitative metagenomics for marine prokaryotes and photosynthetic eukaryotes
Abstract
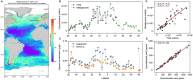
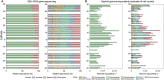
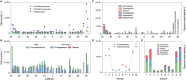
High-throughput sequencing has provided unprecedented insights into microbial biodiversity in marine and other ecosystems. However, most sequencing-based studies report only relative (compositional) rather than absolute abundance, limiting their application in ecological modeling and biogeochemical analyses. Here, we present a metagenomic protocol incorporating genomic internal standards to quantify the absolute abundances of prokaryotes and eukaryotic phytoplankton, which together form the base of the marine food web, in unfractionated seawater. We applied this method to surface waters collected across 50°N to 40°S during the 29th Atlantic Meridional Transect. Using the single-copy recA gene, we estimated an average bacterial abundance of 1.0 × 109 haploid genome equivalents per liter. Leveraging a recent report that the psbO gene is typically single-copy in phytoplankton, we also quantified eukaryotic phytoplankton. Metagenomic estimates closely aligned with flow cytometry data for cyanobacteria (slope = 1.03, Pearson's r = 0.89) and eukaryotic phytoplankton (slope = 0.72, Pearson's r = 0.84). Compared to flow cytometry, taxonomic resolution for nano- and picoeukaryotes was greatly improved. Estimates for diatoms, dinoflagellates, and Trichodesmium were considerably higher than microscopy counts, likely reflecting microscopy undercounts and potential ploidy variation. These findings highlight the value of absolute quantification by metagenomics and offer a robust framework for quantitative assessments in microbial oceanography.
Keywords: absolute quantification; internal standards; metagenomics; phytoplankton.
© The Author(s) 2025. Published by Oxford University Press on behalf of the International Society for Microbial Ecology.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Associated data
LinkOut - more resources
Full Text Sources

