ZBED4: A Prognostic Biomarker and Therapeutic Target in Hepatocellular Carcinoma
- PMID: 40874212
- PMCID: PMC12379359
- DOI: 10.2147/JHC.S546808
ZBED4: A Prognostic Biomarker and Therapeutic Target in Hepatocellular Carcinoma
Abstract
Background: Hepatocellular carcinoma (HCC) is a prevalent lethal cancer that remains challenging to treat. Therefore, investigation of novel targets and therapeutic strategies is essential. The role of ZBED4 in cancer remains unclear.
Methods: Data were sourced from The Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO), International Cancer Genome Consortium (ICGC), and Genomics of Drug Sensitivity in Cancer (GDSC) databases. Various web platforms and R software, have been utilized. Multiplex immunofluorescence was performed on a human HCC tissue microarray.
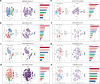
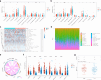
Results: High ZBED4 expression correlates with poor prognosis and immune cell infiltration in multiple cancers. ZBED4 is potentially involved in the regulation of the tumor environment by T cells, with a focus on CD8⁺ T cells. In HCC, tissues with elevated ZBED4 expression exhibit a higher prevalence of Tregs and neutrophils, whereas those with reduced ZBED4 expression show an increased abundance of CD8⁺ T cells, activated CD4⁺ T cells, gamma/delta T cells, and activated natural killer (NK) cells. Elevated ZBED4 expression in HCC patients is associated with a reduced response to immune checkpoint blockade but an improved response to chemotherapy and most targeted therapies. A multi-gene prognostic signature has been developed and confirmed across various HCC cohorts. Multiplex immunofluorescence study demonstrated that ZBED4 was linked to poor prognosis and negatively correlated with CD8⁺ T cell infiltration.
Conclusion: Our research elucidates the role of ZBED4, its strong link to immune infiltration, and its potential as a prognostic and therapeutic biomarker for HCC.
Keywords: ZBED4; hepatocellular carcinoma; immune infiltration; pan-cancer; prognosis; therapy.
Plain language summary
Why was this study done?: Hepatocellular carcinoma (HCC) is a common and aggressive liver cancer with limited treatment options. The ZBED gene family plays roles in cancer progression, but the function of one member, ZBED4, was unknown. This study explored whether ZBED4 could serve as a biomarker to predict patient outcomes or guide therapies for HCC.What did the researchers do?We analyzed data from public cancer databases and patient tissue samples to investigate ZBED4’s role in HCC. We examined: How ZBED4 levels vary in tumors compared to normal tissues.Whether ZBED4 affects immune cells in the tumor environment.Its link to patient survival and response to treatments like chemotherapy and immunotherapy.
What did we find?: High ZBED4 levels were linked to worse survival and advanced cancer stages.Tumors with more ZBED4 had fewer cancer-fighting immune cells (like CD8⁺ T cells) and more immunosuppressive cells (like Tregs).ZBED4 may predict drug sensitivity: patients with high ZBED4 responded better to chemotherapy but poorly to immunotherapy.A 10-gene signature based on ZBED4-related genes helped identify high-risk HCC patients.What do these results mean?ZBED4 could be a useful biomarker to guide treatment decisions in HCC. Targeting ZBED4 might improve outcomes, though further research is needed to confirm this. Future basic and clinical studies should explore whether combining ZBED4-targeted therapies with immune-boosting treatments could enhance their effectiveness.
Key terms explained: Biomarker: A measurable indicator of disease or treatment response.Immunotherapy: Treatments that help the immune system fight cancer.Tumor microenvironment (TME): The surrounding cells and molecules that influence tumor growth.This work opens new avenues of ZBED4 for personalized HCC therapy and highlights its importance in the tumor microenvironment and cancer progression.
© 2025 Ding et al.
Conflict of interest statement
The authors report no conflicts of interest in this work.
Figures







Similar articles
-
Interplay between tumor mutation burden and the tumor microenvironment predicts the prognosis of pan-cancer anti-PD-1/PD-L1 therapy.Front Immunol. 2025 Jul 24;16:1557461. doi: 10.3389/fimmu.2025.1557461. eCollection 2025. Front Immunol. 2025. PMID: 40777041 Free PMC article.
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Systemic treatments for metastatic cutaneous melanoma.Cochrane Database Syst Rev. 2018 Feb 6;2(2):CD011123. doi: 10.1002/14651858.CD011123.pub2. Cochrane Database Syst Rev. 2018. PMID: 29405038 Free PMC article.
-
Senescent fibroblasts secrete CTHRC1 to promote cancer stemness in hepatocellular carcinoma.Cell Commun Signal. 2025 Aug 25;23(1):379. doi: 10.1186/s12964-025-02369-8. Cell Commun Signal. 2025. PMID: 40855439 Free PMC article.
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
References
LinkOut - more resources
Full Text Sources
Research Materials

