BBANsh: a deep learning architecture based on BERT and bilinear attention networks to identify potent shRNA
- PMID: 40874819
- PMCID: PMC12392269
- DOI: 10.1093/bib/bbaf443
BBANsh: a deep learning architecture based on BERT and bilinear attention networks to identify potent shRNA
Abstract
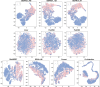
RNA interference (RNAi) is a technique for precisely silencing the expression of specific genes by means of small RNA molecules and is essential in functional genomics. Among the commonly used RNAi molecules, short hairpin RNAs (shRNAs) exhibit advantages over small interfering RNAs, including longer half-life, comparable silencing efficiency, fewer off-target effects, and greater safety. However, traditional screening of potent shRNAs is costly and time-consuming. Advances in big data and artificial intelligence have enabled computational methods to significantly accelerate shRNA design and prediction. In this study, we propose BBANsh, a new shRNA prediction model based on bidirectional encoder representation from transformers (BERT) and bilinear attention network (BAN). We comprehensively evaluate the performance of BBANsh against traditional feature-based models, various feature fusion methods, and existing shRNA prediction models. The BBANsh has achieved an area under the precision-recall curve of 0.951 on five-cross validation and a prediction accuracy of 0.896 on a new external validation set, highlighting its superior predictive performance. Ablation experiments validate the significant contributions of BERT and BAN to model performance. The visualization of internal feature representations intuitively demonstrates the effectiveness of the feature fusion strategy of BBANsh. Furthermore, the attentional analysis reveals that nucleotides near the 5' end have the greatest impact on model predictions, highlighting sequence characteristics of potent shRNAs. Overall, BBANsh provides an efficient and reliable tool for shRNA prediction, which can offer valuable support for researchers in the precise selection and design of shRNA.
Keywords: BERT; bilinear attention network; shRNA prediction.
© The Author(s) 2025. Published by Oxford University Press.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

