Peripherally administered androgen receptor-targeted antisense oligonucleotide rescues spinal pathology in a murine SBMA model
- PMID: 40875460
- PMCID: PMC12578385
- DOI: 10.1172/JCI182955
Peripherally administered androgen receptor-targeted antisense oligonucleotide rescues spinal pathology in a murine SBMA model
Abstract
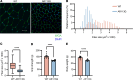
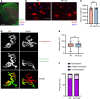
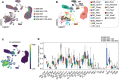
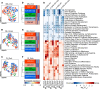
Degeneration of the neuromuscular system is a characteristic feature of spinal and bulbar muscular atrophy (SBMA), a CAG/polyglutamine (polyQ) expansion disorder caused by mutation in the androgen receptor (AR). Using a gene-targeted mouse model of SBMA, AR113Q mice, we demonstrate age-dependent degeneration of the neuromuscular system that initially manifests with muscle weakness and atrophy and progresses to include denervation of neuromuscular junctions and lower motor neuron soma atrophy. Using this model, we tested the hypothesis that therapeutic intervention targeting skeletal muscle during this period of disease progression arrests degeneration of the neuromuscular system. To accomplish this, AR-targeted antisense oligonucleotides were administered subcutaneously to symptomatic AR113Q mice to reduce expression of polyQ AR in peripheral tissues but not in the spinal cord. This intervention rescued muscle atrophy, neuromuscular junction innervation, lower motor neuron soma size, and survival in aged AR113Q mice. Single-nucleus RNA sequencing revealed age-dependent transcriptional changes in the AR113Q spinal cord during disease progression, which were mitigated by peripheral AR gene silencing. Our findings underscore the intricate interplay between peripheral tissues and the central nervous system in SBMA and emphasize the therapeutic effectiveness of peripheral gene knockdown in symptomatic disease.
Keywords: Neurodegeneration; Neuromuscular disease; Neuroscience; Therapeutics.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

