Alterations in ether lipid metabolism in obesity revealed by systems genomics of multi-omics datasets
- PMID: 40875616
- PMCID: PMC12393746
- DOI: 10.1371/journal.pbio.3003349
Alterations in ether lipid metabolism in obesity revealed by systems genomics of multi-omics datasets
Abstract
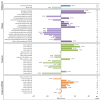
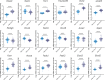
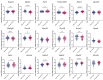
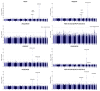
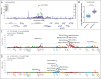
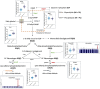
Ratios between two metabolites are sensitive indicators of metabolic changes. Lipidomic profiling studies have revealed that plasma ether lipids, a class of glycero- and glycerophospho-lipids with reported health benefits, are negatively associated with obesity. Here, we utilized lipid ratios as surrogate markers of lipid metabolism to explore the processes underlying the inverse relationship between ether lipid metabolism and obesity. Plasma lipidomics data from two independent human cohorts (n = 10,339 and n = 4,492) were integrated to assess the associations between 82 lipid ratios and obesity-related markers in males and females. Results were externally validated using mouse transcriptomics data from the Hybrid Mouse Diversity Panel (n = 152-227 across 74 strains). Genome-wide association studies using imputed genotypes from a population cohort (n = 4,492) were performed to examine the genetic architecture of the ratios. Findings showed that waist circumference (WC), body mass index, and waist-hip ratio were inversely associated with total plasmalogens relative to total phospholipids in both sexes. Ratios comprising product-substrate pairs positioned either side of enzymes involved in plasmalogen synthesis and degradation showed positive and negative associations with WC, respectively. Branched-chain fatty acids negatively correlated with WC, while omega-6 polyunsaturated fatty acids exhibited differing associations depending on their position within the pathway. Mouse transcriptomics corroborated these results. Genomics data showed strong associations between ratios containing choline-plasmalogens and single-nucleotide polymorphisms in the transmembrane protein 229B (TMEM229B) gene region. This work demonstrates the utility of lipid ratios in understanding lipid metabolism. By applying the ratios to multi-omic datasets, we identified alterations in enzymatic activity and genetic variants likely affecting ether lipid synthesis in obesity that could not have been obtained from lipidomics data alone. Additionally, we characterized a potential role for TMEM229B, offering new perspectives on ether lipid metabolism and regulation.
Copyright: © 2025 Schooneveldt et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
I have read the journal's policy and the authors of this manuscript have the following competing interests: The Baker Heart and Diabetes Institute holds a patent (PCT/AU2020/050742, titled “Composition and Methods of Use”) describing modulation of plasmalogens to improve metabolic health, with PJM listed as the Inventor. This patent has been licensed to Juvenescence Ltd, and a family of related patents have since been filed.
Figures


References
-
- Harwood JL, Michell RH, Murphy DJ, Gurr MI, Frayn KN. Lipids: biochemistry, biotechnology and health. 6th ed. Chichester, West Sussex: John Wiley & Sons; 2016.
-
- Pietiläinen KH, Sysi-Aho M, Rissanen A, Seppänen-Laakso T, Yki-Järvinen H, Kaprio J, et al. Acquired obesity is associated with changes in the serum lipidomic profile independent of genetic effects—a monozygotic twin study. PLoS One. 2007;2(2):e218. doi: 10.1371/journal.pone.0000218 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

