Organoid-based neutralization assays reveal a distinctive profile of SARS-CoV-2 antibodies and recapitulate the real-world efficacy
- PMID: 40875807
- PMCID: PMC12415223
- DOI: 10.1073/pnas.2509616122
Organoid-based neutralization assays reveal a distinctive profile of SARS-CoV-2 antibodies and recapitulate the real-world efficacy
Abstract
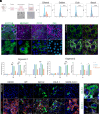
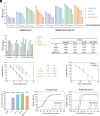
The efficacy of VIR-7831, a class 3 anti-SARS-CoV-2 monoclonal antibody (mAb), was demonstrated repeatedly in clinical trials; yet, reduced neutralization against Omicron variants in cell-line-based neutralization assays led to its withdrawal from clinical use. We developed organoid-based neutralization assays to measure mAb potency. We found that most class 3 mAbs, especially those not blocking receptor-binding domain-ACE2 binding, including VIR-7831, were substantially underestimated in cell-line-based assays. Nasal organoids adequately recapitulated the real-world effectiveness of VIR-7831 because of biologically relevant low ACE2 expression, and exclusively reproduced the in vivo protection of S2 mAbs due to the high TMPRSS2 expression, reminiscent of native human respiratory epithelial cells. Collectively, the robust organoid culture system and biologically relevant expression profiles of ACE2 and TMPRSS2 make nasal organoids present a correlate of in vivo protection of neutralizing mAbs exclusively. The organoid-based neutralization assays, superior to conventional cell-line-based assays, can recapitulate and predict the real-world efficacy of mAbs.
Keywords: SARS-CoV-2; neutralizing antibody; organoid.
Conflict of interest statement
Competing interests statement:C.L., M.C.C., K.Y.Y., and J.Z. are listed as inventors on the patent of nasal organoids (Application Number 63/358,795). Z.W, C.L., Y.Z., L.H., and J.Z. are listed as inventors on the patent of organoid-based neutralization assays. J.Z. is the founder of BiomOrgan Ltd. All other authors declare no competing interests.
Figures



References
-
- Kaku Y., et al. , Virological characteristics of the SARS-CoV-2 KP.3, LB.1, and KP.2.3 variants. Lancet Infect. Dis. 24, e482–e483 (2024). - PubMed
-
- Jian F., et al. , A generalized framework to identify SARS-CoV-2 broadly neutralizing antibodies. bioRxiv [Preprint] (2024). 10.1101/2024.04.16.589454 (Accessed 16 April 2024). - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

