Genome-wide association study identifying immune markers in Qingyuan partridge chickens infected with avian Leukosis virus subgroup J
- PMID: 40876114
- PMCID: PMC12409427
- DOI: 10.1016/j.psj.2025.105732
Genome-wide association study identifying immune markers in Qingyuan partridge chickens infected with avian Leukosis virus subgroup J
Abstract
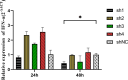
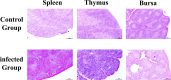
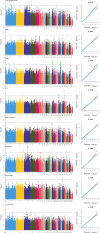
In poultry, Avian Leukosis Virus (ALV) causes high mortality and significant eradication costs, with no effective vaccines, hindering industry development. Improving resistance to ALV-J offers both economic and ecological value. The Qingyuan Partridge Chickens is a high-quality, possesses certain disease resistance capabilities. This study explored resistance molecular markers in Qingyuan Partridge Chickens through cell experiments, viral infection, and genomic resequencing. Results showed that interferon-alpha (IFN-α) expression significantly suppressed the ALV-J duplicated, with notable immune response differences between sexes. Genome-wide association study (GWAS) identified 62 single nucleotide polymorphisms (SNPs) linked to immune responses, cloacal viral positivity, and tissue indices. Selection for the C allele at chromosome 13 positions 8654609-8654610, the G allele at chromosome 9 position 21398106, and the G-T-C haplotype at chromosome 3 positions 59308643-59309366, while selecting against the T and A alleles at chromosome 1 positions 114656880 and 69131730, will effectively enhance immune function and reduce viral susceptibility. Gene Ontology analysis highlighted enrichments in biological processes like glial cell differentiation, cellular components such as side of membrane, and molecular functions like hydrolase activity. Quantitative PCR showed significant expression differences across immune organs in five genes, including IFN-α, interferon β (IFN-β), dual specificity phosphatase 1 (DUSP1), sosondowah ankyrin repeat domain family member C (SOWAHC), and dual specificity phosphatase 10 (DUSP10). These findings provide a genetic basis for breeding ALV-J-resistant Qingyuan Partridge Chickens through marker-assisted selection.
Keywords: Avian Leukosis Virus (ALV); IFN-α; Qingyuan Partridge chickens; genome-wide association study (GWAS).
Copyright © 2025. Published by Elsevier Inc.
Conflict of interest statement
Disclosures No potential conflict of interest was reported by the authors.
Figures

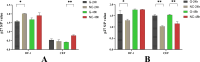






Similar articles
-
Isolation and molecular characteristic of subgroup J avian leukosis virus in Guangxi and Jiangsu provinces of China during 2022-2023.Poult Sci. 2025 Aug;104(8):105272. doi: 10.1016/j.psj.2025.105272. Epub 2025 May 7. Poult Sci. 2025. PMID: 40367569 Free PMC article.
-
Single Amino Acid Residue W33 of tva Receptor Is Critical for Viral Entry and High-Affinity Binding of Avian Leukosis Virus Subgroup K.Viruses. 2025 May 15;17(5):709. doi: 10.3390/v17050709. Viruses. 2025. PMID: 40431720 Free PMC article.
-
Avian leukosis virus p15 interacts with interferon regulatory factor 7 to antagonize the cGAS-STING signaling pathway and promote viral replication.J Virol. 2025 Aug 19;99(8):e0038025. doi: 10.1128/jvi.00380-25. Epub 2025 Jul 24. J Virol. 2025. PMID: 40704898 Free PMC article.
-
Adefovir dipivoxil and pegylated interferon alfa-2a for the treatment of chronic hepatitis B: a systematic review and economic evaluation.Health Technol Assess. 2006 Aug;10(28):iii-iv, xi-xiv, 1-183. doi: 10.3310/hta10280. Health Technol Assess. 2006. PMID: 16904047
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

