Tumor evolution and immune microenvironment dynamics in primary and relapsed mantle cell lymphoma
- PMID: 40876452
- PMCID: PMC12490240
- DOI: 10.1016/j.xcrm.2025.102318
Tumor evolution and immune microenvironment dynamics in primary and relapsed mantle cell lymphoma
Abstract
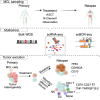
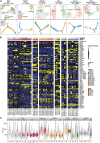
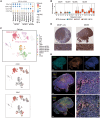
Mantle cell lymphoma (MCL) is a rare but often aggressive type of B cell lymphoma with a high risk of relapse. To explore intratumoral clonal diversity and tumor evolution related to disease relapse, we integrate single-cell RNA and B cell receptor sequencing with whole-genome sequencing in 20 diagnosed/untreated and/or relapsed samples from 11 MCL patients. Our results reveal significant intratumor heterogeneity in MCL already at diagnosis. We further show that the evolutionary paths during disease progression for each patient are unique, where minor clones present at diagnosis may acquire different mutations and copy-number variations and/or migrate to various microenvironments. Despite significant interpatient heterogeneity, recurrent genetic and transcriptomic changes in tumor cells affecting key signaling pathways, along with alterations involved in the tumor microenvironment, are also observed during disease progression. Taken together, our findings elucidate the diverse and dynamic tumor-immune evolution processes associated with disease progression and relapse in MCL.
Keywords: clonal evolution; immunotherapy; mantle cell lymphoma; relapse; scRNA-seq; single-cell RNA sequencing; tumor heterogeneity; tumor microenvironment.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Veloza L., Ribera-Cortada I., Campo E. Mantle cell lymphoma pathology update in the 2016 WHO classification. Ann. Lymphoma. 2019;3:3. doi: 10.21037/aol.2019.03.01. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

