New mechanistic insights into Prp22-mediated exon ligation and mRNA release
- PMID: 40876859
- PMCID: PMC12393901
- DOI: 10.1093/nar/gkaf823
New mechanistic insights into Prp22-mediated exon ligation and mRNA release
Abstract
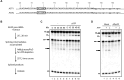
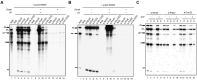
The DExD/H-box RNA helicase Prp22 catalyzes messenger RNA (mRNA) release from the spliceosome, and has also been implicated in proofreading the 3' splice site (3'SS), preventing exon ligation of mutant pre-mRNAs through an ATP-dependent mechanism. However, here we reveal an unexpected role for Prp22 in promoting exon ligation of both wild-type and mutant pre-mRNAs by stabilizing Slu7's association with the spliceosome prior to exon ligation. Notably, ATP binding, rather than hydrolysis, by Prp22 inhibits exon ligation of 3'SS mutant pre-mRNA. Following exon ligation, Prp22-mediated ATP hydrolysis facilitates the dissociation of both Slu7 and mRNA from the spliceosome. Remarkably, Prp22 and Cwc22, which bind the 3'- and 5'-exons respectively, remain associated with the released mRNA, whereas Slu7 and Fyv6 dissociate independently. We propose that Prp22 facilitates exon ligation by stabilizing Slu7 binding, with binding of ATP by Prp22 potentially destabilizing that interaction, thereby weakening contacts between the 5'-exon and the 3'SS to inhibit exon ligation. After exon ligation, Prp22-driven ATP hydrolysis induces a conformational change in Prp8 that disrupts its interdomain interactions, enabling mRNA release through the domain interfaces, with Prp22 and Cwc22 remaining associated with the released mRNA.
Plain language summary
DExD/H-box RNA helicase Prp22 catalyzes the release of mature messenger RNA (mRNA) from the spliceosome and has also been implicated in proofreading the 3′ splice site (3′SS) through an ATP hydrolysis-dependent mechanism. Our study reveals a contrasting, ATP-independent function of Prp22 in promoting exon ligation in both wild-type and 3′SS-mutated pre-mRNAs by stabilizing the binding of step-two factor Slu7. After exon ligation, ATP hydrolysis by Prp22 drives the dissociation of Slu7 and the release of mRNA, with both Prp22 and Cwc22 remaining associated with the released mRNA. We propose that Prp22-mediated ATP hydrolysis induces a conformational change in Prp8 that disrupts its interaction with Slu7 and its interdomain contacts, enabling mRNA release through these Prp8 interfaces upon Slu7 dissociation.
© The Author(s) 2025. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures







References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

