HapA protease targets PAR-1/2 to modulate ERK signalling and reduce cancer cell viability
- PMID: 40877222
- PMCID: PMC12394649
- DOI: 10.1038/s41420-025-02691-7
HapA protease targets PAR-1/2 to modulate ERK signalling and reduce cancer cell viability
Abstract
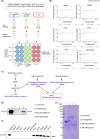
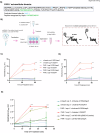
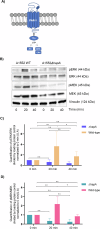
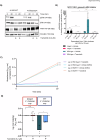
Recent studies reveal that Vibrio cholerae secretes virulence factors impacting host cell viability, though their effects on cancer cells remain unclear. However, the bacterial components and mechanisms influencing cancer cells remain largely unknown. This study investigated the effects of V. cholerae mutants lacking secreted proteins on carcinoma cells. We identified the hemagglutinin zinc-metalloprotease HapA as the main factor reducing cancer cell viability. HapA cleaves protease-activated receptors 1 and 2 on epithelial cancer cells at unique sites, unlike human proteases. This cleavage triggers an early and transient activation of the kinases MEK and ERK. Transient MEK and ERK activation initiates caspase 7, leading to apoptosis and reduced viability in epithelial cancer cells. Our findings underscore the significance of human protease-activated receptors as targets for bacterial protease HapA. Furthermore, we demonstrate that selective cleavage of PAR-1/2 by HapA adjusts MEK-ERK signalling dynamics, suggesting potential new avenues for the development of novel anticancer therapies. Understanding how pathogens like V. cholerae interact with cancer cells sheds light on potential mechanisms underlying cancer progression and suggests new therapeutic targets for cancer treatment.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: AH provides advice to Novartis (clinical trial NeoLetTrib) and receives funding from Menarini pharma company. The other authors declare that there are no conflicts of interest. Ethics approval and consent to participate: We clarify that all experiments were conducted entirely in vitro. All methods were performed in accordance with the relevant guidelines and regulations. The study did not involve human participants, identifiable data or images, patient samples, volunteer-based research, or studies involving live vertebrates. No clinical or drug trials were conducted. Therefore, ethics approval and informed consent requirements do not apply.
Figures
References
-
- Punj V, Bhattacharyya S, Saint-Dic D, Vasu C, Cunningham EA, Graves J, et al. Bacterial cupredoxin azurin as an inducer of apoptosis and regression in human breast cancer. Oncogene. 2004;23:2367–78. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

