DCAF7 recruits USP2 to facilitate hepatocellular carcinoma progression by suppressing clockophagy-induced ferroptosis
- PMID: 40877242
- PMCID: PMC12394690
- DOI: 10.1038/s41419-025-07977-3
DCAF7 recruits USP2 to facilitate hepatocellular carcinoma progression by suppressing clockophagy-induced ferroptosis
Abstract
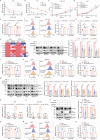
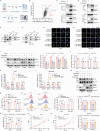
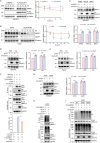
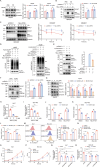
DDB1- and CUL4-associated factor 7 (DCAF7) has recently been identified as a critical regulator of tumorigenesis and a potential modulator of ferroptosis. However, the precise function of DCAF7 in regulating the progression of hepatocellular carcinoma (HCC) ferroptosis remains elusive. In this study, we demonstrate that DCAF7 and the deubiquitinase USP2 are highly expressed in HCC. Genetic ablation of DCAF7 or pharmacological inhibition of USP2 sensitizes HCC to ferroptosis and inhibits HCC progression both in vitro and in vivo. Mechanistically, DCAF7 recruits USP2 to inhibit clockophagy (the selective autophagic degradation of core clock protein BMAL1 mediated through p62/SQSTM1) by reducing BMAL1 K63-linked polyubiquitination. Targeting either DCAF7 or USP2 triggers clockophagy-induced ferroptosis through the HIF1α-SLC7A11 axis in HCC cells. Collectively, our study establishes DCAF7 and USP2 as novel suppressors of clockophagy-induced ferroptosis and reveals the potential therapeutic targets for HCC treatment.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics approval: This study was conducted in compliance with all relevant guidelines and regulations. Full informed consent was obtained from all human participants, and the study protocol was approved by the Ethics Committee of Soochow University (Approval No: SUDA20240522H04). The animal experiments were strictly adhered to protocols approved by the Ethics Committee of Soochow University (Approval No: SUDA20240522A03).
Figures




References
-
- Xu J, Ye Z, Zhuo Q, Gao H, Qin Y, Lou X, et al. MEN1 degradation induced by neddylation and the CUL4B-DCAF7 axis promotes pancreatic neuroendocrine tumor progression. Cancer Res. 2023;83:2226–47. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

