Collagen disorder architecture features are associated with clinical, molecular, genetic factors and survival outcomes in colon cancer
- PMID: 40877439
- PMCID: PMC12394695
- DOI: 10.1038/s41698-025-01098-y
Collagen disorder architecture features are associated with clinical, molecular, genetic factors and survival outcomes in colon cancer
Abstract
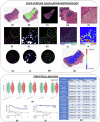
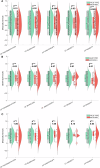
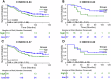
We developed a computational pathology pipeline to extract and analyze collagen disorder architecture (CoDA) features from whole slide images (WSIs) of 2,212 colon cancer (CC) patients across multiple institutions. CoDA features-capturing collagen fragmentation, bundling, anisotropy, density, and rigidity, were evaluated for associations with clinical variables (overall stage, T/N/M stage), molecular classifications (Consensus Molecular Subtypes [CMS1-4]), and genetic mutations (KRAS, BRAF, NRAS) using the Mann-Whitney U test with Bonferroni correction. These analyses revealed significant differences in CoDA feature distributions across multiple subgroups, suggesting that collagen architecture varies meaningfully with tumor stage, molecular subtype, and mutation status.To assess how well CoDA features could distinguish between these subgroups, we implemented a Random Forest classification framework. High mean AUC values (≥0.7) across several variables indicated strong discriminatory performance of CoDA features in separating clinically and biologically distinct groups.For survival analysis, LASSO-Cox models were trained on the PLCO dataset to generate CoDA-based risk scores for overall survival (OS) and disease-free survival (DFS), which were used to stratify patients into high- and low-risk groups in a combined validation dataset (TCGA, UH, and Emory). Kaplan-Meier curves demonstrated significant survival differences across clinical stages, CMS subtypes, and KRAS mutation status. Multivariable Cox proportional hazards models further confirmed the independent prognostic value of CoDA features after adjusting for clinical, molecular, and genetic covariates. These findings highlight that CoDA features are significantly associated with key clinical and molecular characteristics, can distinguish relevant patient subgroups, and offer independent prognostic information, underscoring their potential utility in characterizing the tumor microenvironment and informing risk stratification in CC.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: Dr. Madabhushi is an equity holder at Picture Health, Elucid Bioimaging, and Inspirata Inc. Currently, he serves on the advisory board of Picture Health and SimBioSys. He currently consults Takeda Inc. He also sponsored research agreements with AstraZeneca and Bristol Myers Squibb. His technology has been licensed for Picture Health and Elucid Bioimaging. He is also involved in two different R01 grants from Inspirata, Inc. He also served as a member of the Frederick National Laboratory Advisory Committee. The remaining authors declare no competing interests.
Figures








Similar articles
-
Clinical diagnostic and prognostic value of homocysteine combined with hemoglobin [f (Hcy-Hb)] in cardio-renal syndrome caused by primary acute myocardial infarction.J Transl Med. 2025 Jul 23;23(1):813. doi: 10.1186/s12967-025-06512-4. J Transl Med. 2025. PMID: 40702542 Free PMC article.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Molecular feature-based classification of retroperitoneal liposarcoma: a prospective cohort study.Elife. 2025 May 23;14:RP100887. doi: 10.7554/eLife.100887. Elife. 2025. PMID: 40407808 Free PMC article.
-
Systemic treatments for metastatic cutaneous melanoma.Cochrane Database Syst Rev. 2018 Feb 6;2(2):CD011123. doi: 10.1002/14651858.CD011123.pub2. Cochrane Database Syst Rev. 2018. PMID: 29405038 Free PMC article.
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
References
Grants and funding
- R01 CA216579/CA/NCI NIH HHS/United States
- U01 CA239055/CA/NCI NIH HHS/United States
- R01 CA268287/CA/NCI NIH HHS/United States
- R01 CA249992/CA/NCI NIH HHS/United States
- R01 CA220581/CA/NCI NIH HHS/United States
- R01 CA202752/CA/NCI NIH HHS/United States
- R01 CA208236/CA/NCI NIH HHS/United States
- U01 CA248226/CA/NCI NIH HHS/United States
- I01 BX004121/BX/BLRD VA/United States
- R43 EB028736/EB/NIBIB NIH HHS/United States
- U01 CA269181/CA/NCI NIH HHS/United States
- R01 CA257612/CA/NCI NIH HHS/United States
- U54 CA254566/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

