MS4A6A/Ms4a6d deficiency disrupts neuroprotective microglia functions and promotes inflammation in Alzheimer's disease model
- PMID: 40877951
- PMCID: PMC12395693
- DOI: 10.1186/s13024-025-00887-0
MS4A6A/Ms4a6d deficiency disrupts neuroprotective microglia functions and promotes inflammation in Alzheimer's disease model
Abstract
Background: Alzheimer's disease (AD) is the most common type of dementia. Genetic polymorphisms are associated with altered risks of AD onset, pointing to biological processes and potential targets for interventions. Consistent with the important roles of microglia in AD development, genetic mutations of several genes expressed on microglia have been identified as risks for AD. Emerging evidences indicate that the expression of a microglia-specific gene MS4A6A is thought to be associated with AD, since AD patients show upregulation of MS4A6A, and its levels correlate with the severity of clinical neuropathology. However, the mechanism linking MS4A6A and AD has not been experimentally studied.
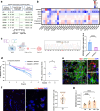
Methods: We performed a meta genome-wide association analysis with 734,121 subjects to examine the associations between polymorphisms of MS4A6A with AD risks. In addition, we analyzed the correlation between MS4A6A and AD-related cerebrospinal fluid biomarkers from our own cohort. Furthermore, we for the first time generated a Ms4a6d deficient APP/PS1 model, and systematically examined pathological changes using high-resolution microscopy, biochemistry, and behavioral analysis.
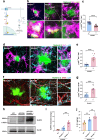
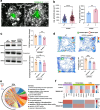
Results: We identified several new mutations of MS4A6A with altered AD risks, and discovered specific correlation for some of them with the amount of β-amyloid in cerebrospinal fluid. Protective variant of MS4A6A is associated with elevated expression of the gene. Deficient Ms4a6d led to reduced amyloid clearance in the brain. Immunostaining from postmortem AD patients brain revealed selective expression of MS4A6A in microglia. In APP/PS1 mice lacking Ms4a6d, microglia showed markedly diminished envelopment and phagocytosis of amyloid, leading to increased plaque burden, less compact structure, and more severe synaptic damage. Importantly, Ms4a6d deficiency markedly exacerbated inflammatory responses in both microglia and astrocytes by disinhibiting NF-κB signaling. Overexpressing MS4A6A in human microglia cell line promoted gene expression related to plaque-associated responses and diminished inflammation signatures.
Conclusions: Our findings reveal that Ms4a6d deficiency suppresses neuroprotection and worsens neuroinflammation. Sufficient Ms4a6d maybe beneficial for boosting amyloid-related responses and suppressing inflammation in microglia, making it superior than previously reported candidates for microglia modulation. Thus, the elevated MS4A6A levels in AD are likely compensatory and boosting MS4A6A could be an effective treatment.
Keywords: Alzheimer’s disease; MS4A6A; Microglia; Ms4a6d; Neuroinflammation.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The CABLE study was carried out following the Declaration of Helsinki and the Qingdao Municipal Hospital's Institutional Ethics Committees approved the study's protocol. The current analyses were conducted under UK Biobank application number 19542. All of the experiments comply with the animal study protocol approved by the Fudan University. The consent for human body and organ donation (code number: 00303) was carefully signed by the donor. Consent for publication: Not applicable. Competing interests: The authors declare that they have no competing interests.
Figures


References
-
- van Dyck CH, Swanson CJ, Aisen P, Bateman RJ, Chen C, Gee M, et al. Lecanemab in early Alzheimer’s disease. N Engl J Med. 2023;388:9–21. - PubMed
-
- Sims R, Hill M, Williams J. The multiplex model of the genetics of Alzheimer’s disease. Nat Neurosci. 2020;23:311–22. - PubMed
-
- Migliore L, Coppedè F. Gene-environment interactions in Alzheimer disease: the emerging role of epigenetics. Nat Rev Neurol. 2022;18:643–60. - PubMed
MeSH terms
Substances
Grants and funding
- 82071201/National Natural Science Foundation of China
- 32371036/National Natural Science Foundation of China
- 2022ZD0211600/Science and Technology Innovation 2030 Major Projects
- 2023SHZDZX02/Shanghai Municipal Science and Technology Major Project
- 2022JC01/Shanghai Municipal Health Commission Emerging Interdisciplinary Research Project
LinkOut - more resources
Full Text Sources
Medical

